- Title
-
Noonan and LEOPARD syndrome Shp2 variants induce heart displacement defects in zebrafish
- Authors
- Bonetti, M., Paardekooper Overman, J., Tessadori, F., Noël, E., Bakkers, J., den Hertog, J.
- Source
- Full text @ Development
|
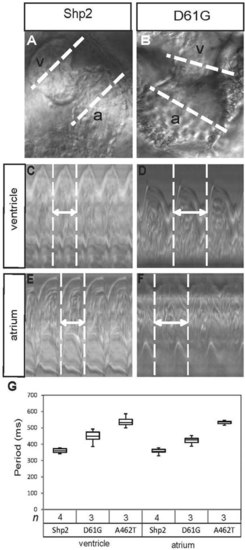
Impaired cardiac function in embryos expressing Shp2-D61G and Shp2-A462T. (A,B) The hearts of WT-Shp2- (Shp2) and Shp2-D61Gexpressing zebrafish embryos were imaged by high-speed video recording microscopy at 2 dpf. White dotted lines through the atrium (a) and the ventricle (v) are indicated. (C-F) Ventricular (C,D) and atrial (E,F) kymographs from 2 dpf embryonic hearts. Note the longer period of the Shp2-D61G-expressing heart, compared with the WT-Shp2-expressing heart (double-headed arrow and white dotted vertical lines). (G) Quantitative analysis of the heart period in the ventricle or atrium of WT-Shp2, Shp2-D61G and Shp2-A462T expressing embryos (n indicates number). Whisker plots are depicted. |
|
Heart defects in embryos expressing Shp2-D61G and Shp2-A462T. Non-injected control embryos (NIC) and embryos injected at the one-cell stage with mRNA encoding WT-Shp2 (Shp2), Shp2-D61G or Shp2-A462T were fixed at 48 hpf and in situ hybridization was performed using probes for the myosin genes myl7 (A-D, cardiomyocytes), vmhc (E-H, ventricle) and amhc (I-L, atrium), and for has2 (M-P, endocardial cushions). Representative pictures are shown and the number of embryos showing this pattern/total number of embryos is indicated. The outline of the heart is indicated in M-P. EXPRESSION / LABELING:
|
|
Randomized heart displacement in response to expression of Shp2-D61G and Shp2-A462T. Embryos were injected at the one-cell stage with mRNA encoding WT-Shp2, Shp2-D61G or Shp2-A462T and fixed at the 16-somite stage, the 22-somite stage or at 28hpf. In situ hybridization was carried out using a myl7-specific probe to mark cardiomyocytes. (A-H) The number of embryos displaying the depicted pattern/total number of embryos is indicated. (I-K) Representative pictures are shown displaying heart displacement to the left (I), middle (J) and right (K). (L) Percentages of left, middle and right cardiac displacement are depicted. The number of embryos (n) is indicated. EXPRESSION / LABELING:
|
|
Impaired cardiomyocyte migration in embryos expressing Shp2-D61G and Shp2-A462T. Embryos from the tg(myl7:GFP) line were injected with mRNA encoding WT-Shp2, Shp2-D61G or Shp2-A462T lacking eGFP-peptide 2A sequences to allow imaging of GFP-positive cardiomyocytes from the 22-somite stage onwards. (A-F) Representative individual GFP-positive cells were color-coded according to their location within the cardiac field at the 22-somite stage and tracked over a 200 min period. Dorsal view with anterior towards the top. Schematic representations of the embryos on the left indicate the position and shape of the heart in green. (A-C) WT-Shp2; (D-F) Shp2-A462T. (G) Quantification of the total speed of posterior (P) and anterior (A) cardiac progenitor cells. WT-Shp2- expressing embryos displayed normal leftward heart displacement; embryos expressing Shp2-D61G and Shp2-A462T that did not display normal heart displacement were selected for further analysis. (H) Clockwise cardiomyocyte rotation was determined of the same embryos as in G. Rotation was quantified as depicted in I. (G,H) Averages are indicated and error bars indicate s.e.m.; n indicates number of embryos. Statistical significance was determined using Student’s t-test: **P<0.01 and ***P<0.001. |
|
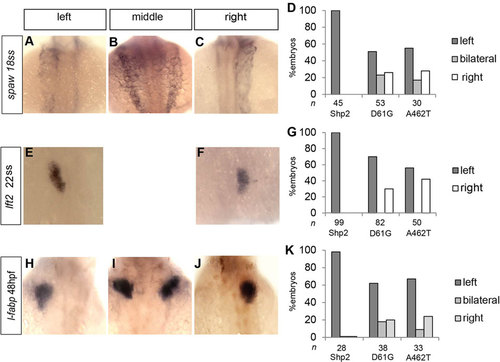
Expression of Shp2-D61G- and Shp2-A462T-induced L/R asymmetry defects. Embryos were injected at the one-cell stage with mRNA encoding WT-Shp2 (Shp2), Shp2-D61G or Shp2-A462T and fixed at the stage indicated. In situ hybridization was carried out using probes specific for spaw (A-D, southpaw), lft2 (E-G, lefty2) and fabp (H-K, fatty acid binding protein, marking the liver). Representative pictures are shown of Shp2-D61G-expressing embryos. lft2 expression was observed only on the left side or on the right side. Organ asymmetry as assessed using the different markers was scored for embryos injected with WT-Shp2, Shp2-D61G and Shp2-A462T. Percentages of left, middle/bilateral and right expression of the markers are depicted. The number of embryos (n) is indicated. |
|
Early treatment with the MEK-inhibitor CI-1040 rescued leftward heart displacement and cilia length in embryos expressing NS and LS Shp2 variants. Embryos were injected with mRNA encoding WT-Shp2 (Shp2), Shp2-D61G and Shp2-A462T at the one-cell stage. (A-F) Representative images of bud stage embryos are depicted. The shape of the NS- and LS-Shp2-expressing embryos is restored by treatment with 0.25µM CI-1040 at 4.5hpf for 1h. (G) The ratio between the length of the major and minor axes was determined as quantitative measure of the epiboly defects in NS- and LS-Shp2-expressing embryos. Averages are depicted with error bars indicating s.e.m. Statistical significance was determined using Student′s t-test (**P<0.01; ***P<0.001). (H) Embryos were treated with MEK inhibitor CI-1040 (0.25µM) for 1h at 4.5hpf or mock-treated with DMSO and lysed at 10hpf. Immunoblots of the zebrafish lysates were stained using antibodies specific for pErk and Erk. The experiment was repeated three times and representative blots are depicted here. (I-K) Embryos were mock treated with DMSO (I) or with CI-1040 for 1h at 4.5hpf (J) or at 10hpf (K). Heart displacement was determined in embryos following in situ hybridization with a myl7-specific probe as in Fig. 4. Histograms represent the percentage of embryos with heart displacement to the left, middle or right. The total number of embryos (n) is indicated. |
|
EGFP-Peptide 2A-Shp2 fusions induced defects in zebrafish embryos. (A) Schematic representation of eGFP-Peptide 2A-Shp2 with D61G, T73I (NS) and A462T, G465A (LS) mutations indicated. The full length fusion protein is produced and is then cleaved autoproteolytically, resulting in eGFP and (mutant) Shp2. (B) One- or two-cell stage embryos were injected with in vitro transcribed mRNAs encoding the indicated proteins. Immunoblotting analysis reveals that autoproteolytic cleavage of the fusion proteins is highly efficient in zebrafish embryos. The injected embryos were lyzed at 10 hpf, proteins were separated on SDS-polyacrylamide gels, blotted and probed with antibodies specific for Shp2, GFP and β-actin as a loading control. (C-D) Two representative injected embryos (D61G and A462T) show craniofacial defects (red asterisks), a decrease in body length and cardiac edema (blue asterisks). These defects are absent in WT-Shp2 injected control embryos. Injection efficiency was checked by fluorescence microscopy with a GFP filter. |
|
Heart looping defects in embryos expressing Shp2-T73I (NS) and Shp2- G465A (LS). Non-injected control embryos (NIC) and embryos injected at the one-cell stage with mRNA encoding WT-Shp2, Shp2-T73I (NS) or Shp2-G465A (LS) were fixed at 48 hpf and in situ hybridization was performed using probes for the myosin genes myl7 (A-D, cardiomyocytes), vmhc (E-H, ventricle), amhc (I-L, atrium), and for has2 (M-P, endocardial cushions). Representative pictures are shown and the number of embryos showing this pattern as well as the total number of embryos that were analyzed is indicated in the bottom right corner of each panel. The outline of the heart is indicated with a dashed line in panels M-P. |
|
Midline structures are intact in NS and LS Shp2-variants. (A-C) Brightfield images of (A) WT-Shp2 control, (B) Shp2-D61G and (C) Shp2-A462T expressing embryos, showing normal notochord formation. (D-I) In situ hybridization analysis showed contiguous shh expression in the floorplate. (D-F) Composite pictures of lateral views. (G-I) Dorsal views. The number of embryos showing the depicted expression pattern/ total number of embryos is: WT-Shp2, n=20/20; Shp2-D61G, n=30/30; Shp2-A462T, n=30/30. |