- Title
-
Growth hormone receptors in zebrafish (Danio rerio): adult and embryonic expression patterns
- Authors
- Di Prinzio, C.M., Botta, P.E., Barriga, E.H., Ríos, E.A., Reyes, A.E., and Arranz, S.E.
- Source
- Full text @ Gene Expr. Patterns
|
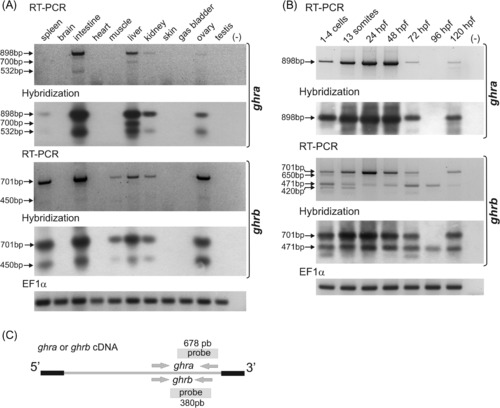
Expression patterns of zebrafish ghra and ghrb in adult tissues (A) and during embryonic development (B), studied by RT-PCR and Southern blotting. EF1α transcripts were amplified as control. PCR with no template were carried out as negative controls (-). The full-length (expected) and the shorter amplicons are indicated by arrows (A and B). Sizes of the amplicons are: 898, 532, and 700 bp for ghra and 701, 420, 471 and 650 bp for ghrb. (C) Positions of primer pairs used in RT-PCR and probes used in Southern blotting for ghra and ghrb cDNAs. |
|
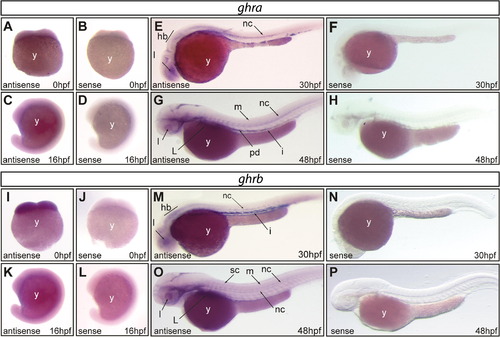
Whole-mount in situ hybridization of zebrafish embryos. Expression of ghra and ghrb mRNAs analyzed at the indicated stages of zebrafish embryos with antisense (A, C, E,G, I, K, M and O) and sense (control) probes (B, D, F, H, J, L, N and P). Ghra and ghrb are expressed ubiquitously from 0 hpf onwards (A and I), indicating they are maternal transcripts. At 16 hpf (C and K) expression continue to be ubiquitous and at 30 hpf several structures are differentially stained. Ghra expression was observed in the lens, hindbrain and the notochord (E), and ghrb transcripts were localized in the hindbrain, lens, notochord and intestine (M). At 48 hpf ghra continued to be detected in the notochord and lens and was detected in the liver, myotomes and pronephric ducts (G). Ghrb was detected in the lens, liver, myotomes, notochord and spinal cord (O). An intense positive signal in the yolk cell was observed for both receptors from 16 hpf onwards (C, E, G and K, M, O). We did not detect positive signals when the embryos and larvae were incubated with the sense probes (B, D, F, H, J, L, N and P). Abbreviations: l, lens; hb, hindbrain; nc, notochord; y, yolk cell; m, myotome; pd, pronephric duct; sc, spinal cord; L, liver. |
|
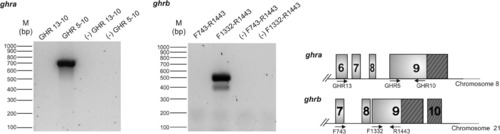
Genomic PCRs of ghra and ghrb. Primer pairs used match in different exons (GHR13–10, F743–R1443) or in exon 9 (GHR5–10, F1332–R1443) as shown in the scheme (right). PCR with no template were carried out as negative controls (-), for each primer pair. Only the expected amplification product was obtained for ghra and two amplification products were obtained for ghrb. No product was obtained with primer pairs that match in different exons in both cases. The diagram on the right shows the approximate position of the primers used in the PCR. |
Reprinted from Gene expression patterns : GEP, 10(4-5), Di Prinzio, C.M., Botta, P.E., Barriga, E.H., Ríos, E.A., Reyes, A.E., and Arranz, S.E., Growth hormone receptors in zebrafish (Danio rerio): adult and embryonic expression patterns, 214-225, Copyright (2010) with permission from Elsevier. Full text @ Gene Expr. Patterns