- Title
-
The Paf1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish
- Authors
- Nguyen, C.T., Langenbacher, A., Hsieh, M., and Chen, J.N.
- Source
- Full text @ Dev. Biol.
|
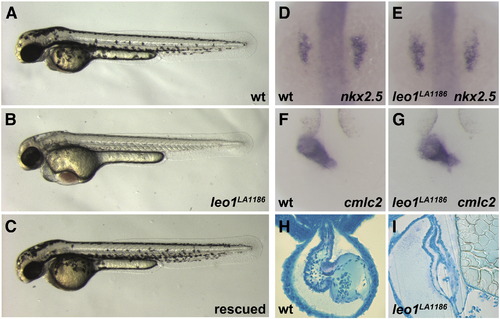
Loss of Leo1 function results in cardiac abnormalities and absence of pigmentation (A, B, C) Lateral view of a two-day-old wild-type embryo (A), a leo1LA1186 mutant (B) and a leo1LA1186 mutant injected with wild-type leo1 mRNA (C). (D, E, F, G) Dorsal view. Expression patterns of cardiac markers, nkx2.5 (D, E) and cmlc2 (F, G), are indistinguishable between wild-type siblings (D, F) and leo1LA1186 mutants (E, G) at the 10 somite stage and 24 h post fertilization, respectively. (H, I) Histological sections. In contrast to wild-type hearts (H), the cardiac chambers of leo1LA1186 mutants are collapsed by 2 dpf (I). o, outflow tract; v, ventricle; a, atrium; sv, sinus venosus. EXPRESSION / LABELING:
PHENOTYPE:
|
|
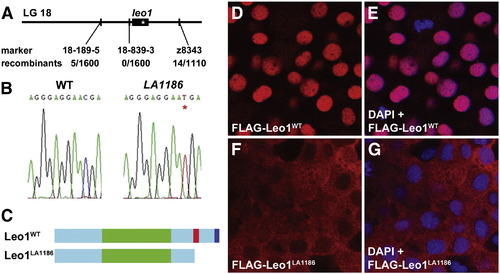
Positional cloning of Leo1LA1186. (A) LA1186 was mapped to linkage group 18 between markers z8343 and 18-189-5. No recombinants were detected between LA1186 and the custom marker 18-839-3 from 1600 meioses. Asterisk marks the position of the nonsense mutation. (B) Sequencing of the leo1 locus in wildtype and LA1186 embryos identified a C to T transition, resulting in a premature stop codon (*). (C) Schematic diagram of the molecular structure of leo1. Zebrafish Leo1WT contains a Leo1 domain (green), nuclear localization sequence (red), and an aspartic acid-rich tail (dark blue). The Leo1LA1186 protein is truncated prior to the nuclear localization sequence. (D, E, F, G) Cellular localization of Leo1WT and Leo1LA1186. FLAG-tagged Leo1WT (D) colocalized with DAPI (E) in the nucleus. FLAG-tagged Leo1LA1186 was detected mostly in the cytoplasm (F, G). FLAG, red; DAPI, blue. EXPRESSION / LABELING:
|
|
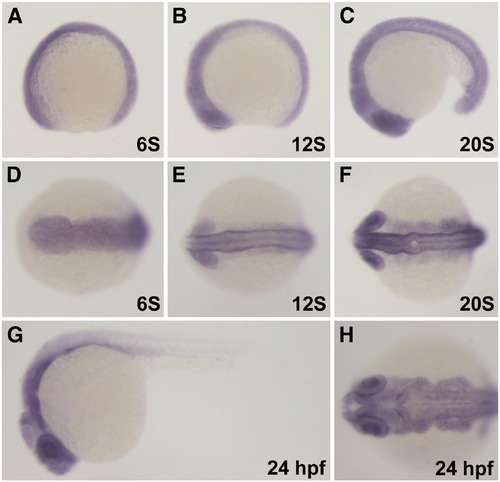
In situ analysis of leo1 mRNA expression. (A–F) Leo1 transcripts are detected in many tissues beginning from the 6S to the 20S stage. (G–H) At 24 hpf, leo1 is strongly expressed in the brain, pharyngeal arches and eyes and tapers posteriorly in the somites. (A, B, C, E, G) Lateral view. (D, E, F, H) Anterior dorsal view. |
|
Cardiac differentiation phenotypes of leo1LA1186. Normally, vmhc expression is restricted in the ventricle in wild-type embryos (A), but is expanded into the atrium in leo1LA1186 mutants (B). Expression of amhc, however, is restricted properly to the atrial chamber in both wild-type (C) and leo1LA1186 mutant embryos (D). Notch1b transcripts are detected in the endocardium at the AV boundary of wild-type embryos (E), but are absent in leo1LA1186 mutants (F). Expression of bmp4 (G), versican (I), and wnt2bb (K) are detected in the myocardium of the AV boundary in wild-type embryos. Both bmp4 (H) and versican (J) transcripts are no longer restricted in leo1LA1186 mutants, while wnt2bb is not detected in mutant embryos (L). Dashed lines (in A–D) and arrows (in E, G, I, K) mark the boundary of atrium and ventricle. EXPRESSION / LABELING:
|
|
Pigmentation phenotypes of leo1LA1186. (A, B) Gch is expressed in xanthophores of the head and trunk (A). Expression levels of gch are reduced in leo1LA1186 mutants (B). (C, D) Dct, a marker for differentiated melanophores, is severely reduced in leo1LA1186 mutants (D) compared to wild-type siblings (C). (E, F) Mitfa transcripts are detected in early melanoblasts along the head and trunk (E). Leo1LA1186 mutants display a reduction in mitfa expression (F). |
|
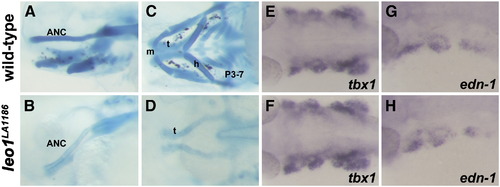
Craniofacial phenotypes of leo1LA1186. (A, B, C, D) Alcian blue staining of wild-type (A, C) and leo1LA1186 embryos (B, D) at 3 days post fertilization. Leo1LA1186 exhibit severe craniofacial defects including absence of the ethmoid plate from the anterior neurocranium and complete loss of pharyngeal arches. (A, B) Lateral view. (C, D) Ventral view. ANC, anterior neurocranium; t, trabecular cranii; m, mandibular arch; h, hyoid arch; P3–7, pharyngeal arches 3–7. (E, F, G, H) Tbx1 and endothelin-1 (edn-1) expression patterns in the pharyngeal arch epithelium and mesenchymal core are indistinguishable between wild-type embryos (E, G) and leo1LA1186 mutants (F, H) at 27 hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
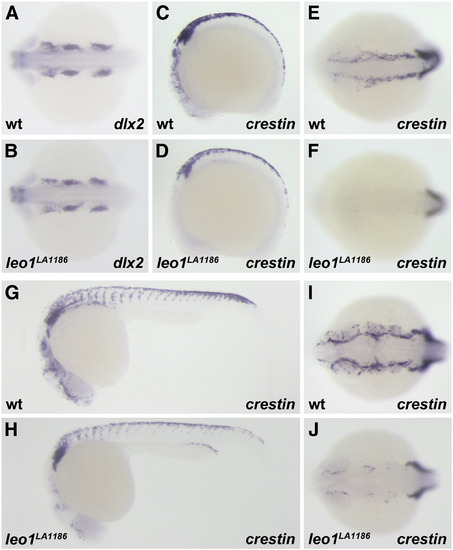
Development of neural crest cells is dependent upon Leo1 function. (A, B) Dorsal anterior view of dlx2 expression in pharyngeal arch neural crest at the 18S stage. The expression pattern of dlx2 is normal in wild-type (A) and leo1LA1186 embryos (B). (C–J) Pan-neural crest marker, crestin, at the 12 somite stage (C–F) and 24hpf (G–J). Crestin expression is detected in anterior and trunk neural crest of wild-type embryos (C, E, G, I), but is severely reduced in the region anterior to the otic vesicle of leo1LA1186 mutants (D, F, H, J). (C, D, G, H) Lateral view. (E, F, I, J) Dorsal anterior view. EXPRESSION / LABELING:
PHENOTYPE:
|
|
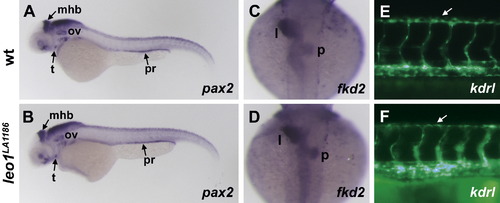
Leo1LA1186 mutants do not have general developmental delay. (A, B) pax2 expression is detected in the midbrain–hindbrain boundary (mhb), thyroid (t), otic vesicle (ov), and pronephric ducts (pr) of wild-type embryos (A) at 50 hpf. Leo1LA1186 mutants have normal expression of pax2 in all of these structures at 50 hpf (B), indicating that the timely organogenesis of the brain, thyroid, otic vesicle, and pronephric ducts does not require Leo1 activity. (C, D) At 50 hpf, both wild-type (C) and leo1LA1186 mutant (D) embryos have a leftward-looped gut, liver (l), and pancreas (p) labeled by fkd2 staining, demonstrating that the morphogenesis of the gut is not developmentally delayed upon loss of Leo1 activity. (E, F) By 50 hpf, the primary intersegmental vessels have branched dorsally to form the dorsal longitudinal anastomotic vessel (arrows) in both wild-type (E) and leo1LA1186 mutant (F) embryos, suggesting that Leo1 activity is not essential for patterning of the trunk vasculature within a normal developmental timeframe. Vessels in E and F are visualized by fluorescence microscopy of Tg(kdrl:GFP)la116 transgenic embryos. |
|
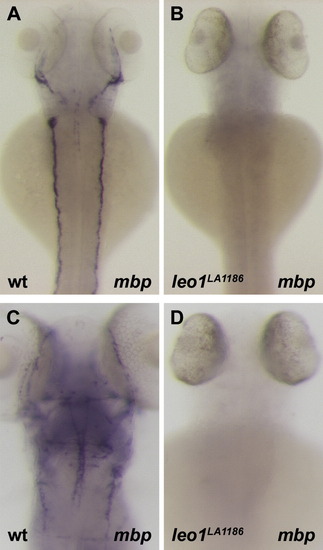
Absence of glial cells in leo1LA1186. (A, C) Dorsal view of mbp expression in 3 dpf (A) and 5dpf (C) wild-type embryos. Expression of mbp was detected in the glial cells of the head and trunk. (B, D) Dorsal view of mbp expression in 3 dpf (B) and 5dpf (D) leo1LA1186 embryos. No expression was detected, indicating a loss of all glial cells. EXPRESSION / LABELING:
PHENOTYPE:
|
|
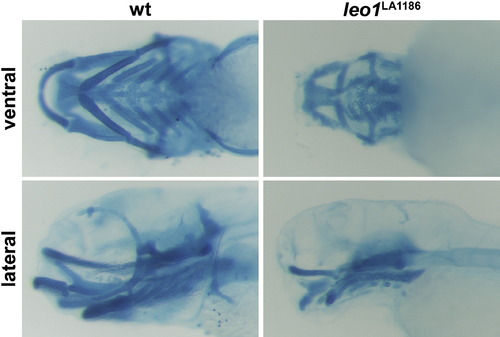
Alcian blue staining. The cartilage component of the craniofacial structure is severely defective in 5-day-old leo1LA1186 mutant embryos. Specifically, the size of each skeletal component is drastically reduced compared to wild-type. |
|
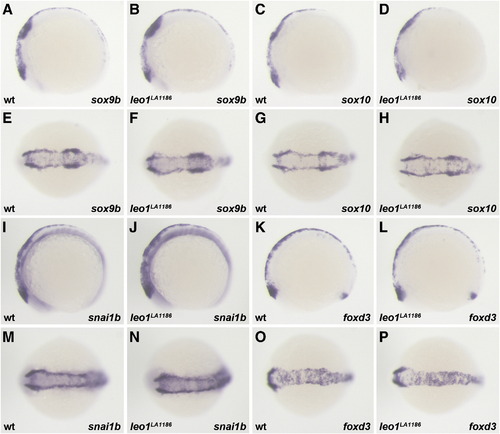
Neural crest specification is unaffected by loss of Leo1 activity. (A–P) Sox9b, sox10, snai1b, and foxd3 are expressed in pre-migratory neural crest at the 10S stage. Their expression patterns are unchanged between wild-type (A, C, E, G, I, K, M, O; wt, wild-type) and leo1LA1186 embryos (B, D, F, H, J, L, N, P). (A–D, I–L) Lateral view. (E–H, M–P) Dorsal view. |

Unillustrated author statements |
Reprinted from Developmental Biology, 341(1), Nguyen, C.T., Langenbacher, A., Hsieh, M., and Chen, J.N., The Paf1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish, 167-175, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.