- Title
-
ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish
- Authors
- Qu, X., Jia, H., Garrity, D.M., Tompkins, K., Batts, L., Appel, B., Zhong, T.P., and Baldwin, H.S.
- Source
- Full text @ Dev. Biol.
|
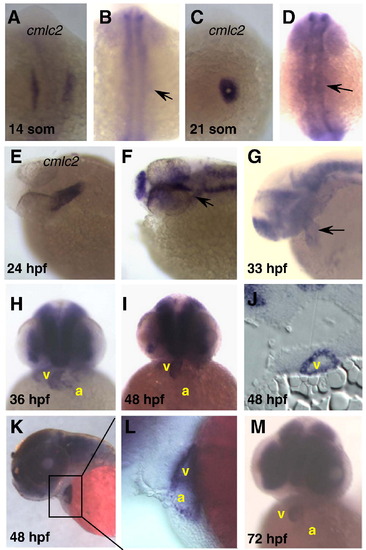
Whole mount zebrafish embryos processed for in situ hybridization using cmlc2 (A, C, E) or ndrg4 (B, D, F–M) specific anti-sense RNA probes. cmlc2 is expressed in the bilateral cardiac primordia at the 14-somite stage (A), in the cardiac cone at the 21-somite stage (C) and in the heart tube at the 24 hpf (E). The first significant indication of ndrg4 cardiac expression occurs in the heart tube (arrow) at 24 hpf (F) and at 33 hpf (G). ndrg4 is robustly expressed in both atrium and ventricle at 36 hpf (H), 48 hpf (I–L) and 72 hpf (M) with stronger signal in the ventricle. (J) Transverse section showing strong expression of ndrg4 in the ventricle at 48 hpf. (L) Higher magnification of the indicated heart region in panel K. (A–D) Dorsal views with anterior towards the top; (E–G, K, L) lateral views; (H, I, M) frontal views with anterior towards the top. a, atrium; v, ventricle. Arrow indicates heart or heart region. EXPRESSION / LABELING:
|
|
Expression of ndrg4 during zebrafish nervous system development by whole mount in situ hybridization. (A) ndrg4 was observed for the first time in the CNS at the 10-somite stage. (B) By 21-somites, ndrg4 is expressed in telencephalon, ventral diencephalon and spinal chord. (C–E) similar expression patterns of ndrg4 in CNS and sensory system at 22–27 hpf. Expression of ndrg4 in nervous system is increased at 33 hpf (F), 48 hpf (G) and 72 hpf (I). (H) Transverse section at 48 hpf showing the strong signal in the retina of the eyes with restriction of expression from the retina proliferative zone (rpz). (A, C–E,G) Dorsal views with anterior towards the left; (B, F, H), lateral views. allg, anterior lateral line ganglion; ce, cerebellum; di, diencephalons; pllg, posterior lateral line ganglion; rm, rhombomeres; teg, tegmentum; tel, telencephalon; trg, trigeminal ganglion. EXPRESSION / LABELING:
|
|
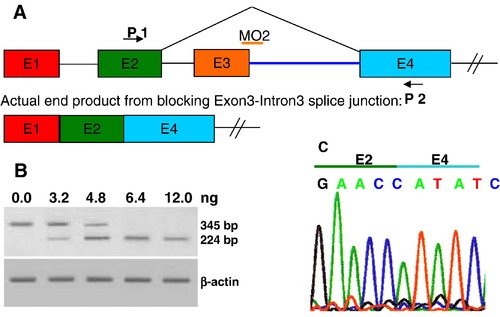
Validation of ndrg4-MO2 (knockdown by splice-blocking). (A) Schematic of the strategy to target the exon3–intron3 boundary with ndrg4-MO2. Arrows represent primers used in the RT-PCR to test the efficiency of ndrg4 splice-blocking MO2 and the orange bar represents the ndrg4-MO2. (B) The RT-PCR was performed on wild-type embryos and embryos injected with 3.2–12.0 ng of the ndrg4-MO2. The ndrg4-MO2 efficiently blocks ndrg4 mRNA splicing by exon3 deletion in a dose-dependent manner. Bands of 345 bp and 224 bp represent wildtype and exon3-deleted ndrg4 mRNA, respectively. (C) Sequencing of the 224 bp band reveals direct connection of exon 2 and exon 4. EXPRESSION / LABELING:
|
|
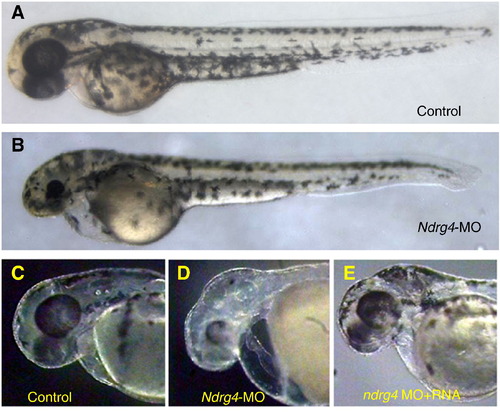
ndrg4 morphant phenotypes. Lateral views at 48 hpf. (A, B) The overall morphology of an uninjected (A) and ndrg4-MO2-injected (B) embryos. Compared to its sibling (C), ndrg4-MO2 injected embryo (D) displays pericardial edema, dilated atrium, unlooped, stretched heart and head defects (smaller eyes and prominent edema in the hindbrain). (E) Co-injection of ndrg4 mRNA with ndrg4-MO2 significantly rescued the cardiac, eye and brain phenotype induced by ndrg4 inhibition. PHENOTYPE:
|
|
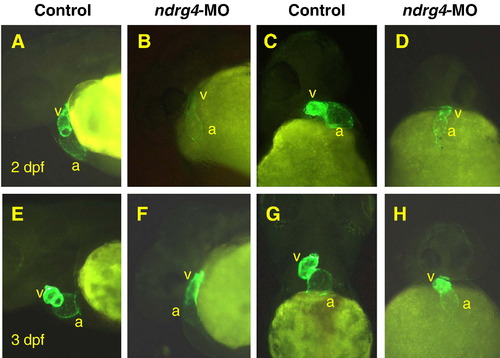
Attenuation of Ndrg4 results in early looping abnormalities. The cmlc2-driven GFP is expressed in cardiomyocytes of transgenic Tg(cmlc2:gfp) embryos. At 2 dpf (A–D) and 3 dpf (E–H), compared to the clearly looped heart of uninjected Tg(cmlc2:gfp) embryos (A, C, E, G), the heart of ndrg4 morphant (B, D, F, H) remained central and linear in a tubular structure. (A, B, E, F), Lateral views with anterior towards the left; (C, D, G, H), frontal views with anterior towards the top. a, atrium; v, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
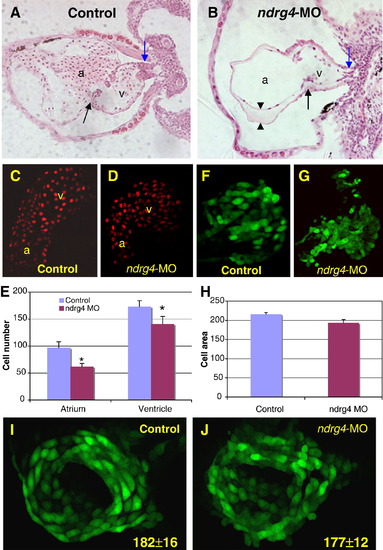
Attenuation of Ndrg4 results in smaller cardiac chambers and a reduction in the number of cardiomyocytes. Transverse sections of 3 dpf wild-type (A) and ndrg4 morphant (B) heart. Outflow tract (blue arrows), AV junction (black arrows) and the enlarged space (arrowheads) between myocardium and endocardium in the dilated atrium are indicated. Note only a few blood cells in the MO treated heart. Confocal images of the heart of control (C) and ndrg4-MO2 injected (D) Tg(cmlc2:DsRed2-nuc) embryos at 2 dpf. (E) Comparison of the numbers of cardiomyocytes in un-injected and ndrg4-MO2 injected embryos at 2 dpf (Data are from 5 control and 5 injected embryos and are expressed as means ± S.D. *P < 0.05). Confocal images of the heart of control (F) and ndrg4-MO2 injected (G) Tg(cmlc2:gfp) embryos at 2 dpf. (H) Comparison of the size of cardiomyocytes in control and ndrg4-MO2 injected embryos at 2 dpf. There was no change in myocardial cell number between control (I) and ndrg4-MO2 (J) injected Tg(cmlc2:gfp) embryos at 24 hpf based on the number of GFP positive cells in the heart: 182 ± 16 vs. 177 ± 12, respectively (Data are from 5 embryos for each group and are expressed as means ± S.D.) a, atrium; v, ventricle; control, mismatch-MO. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Attenuation of Ndrg4 induces a reduction in cell proliferation of the cardiomyocytes. Although no significant difference in the number of BrdU positive cells between control (A) and ndrg4-MO2 treated (B) embryonic hearts at 31 hpf has been detected, ndrg4-MO2 injected embryos (D) display lower number of proliferating cells than control (C) in the heart at 48 hpf. All images represent reconstructions of confocal Z-stack sections imaged on whole hearts. Green, anti-GFP antibody; Red, anti-BrdU antibody. (E) Histogram showing proliferation index (the average of the total number of BrdU positive cells in per 100 cardiomyocytes) in the heart of 48 hpf embryos: control, 13.5 ± 3.9; ndrg4-MO2, 7.8 ± 2.1 (*P < 0.01). |
|
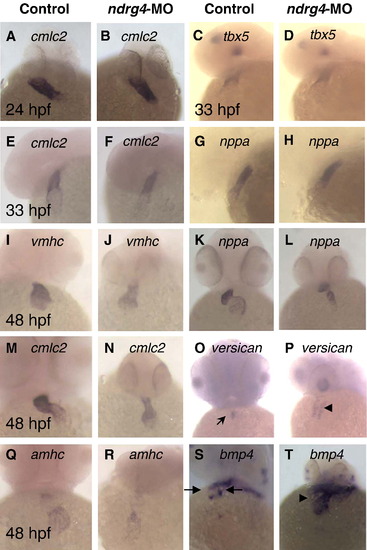
Effect of ndrg4 knock-down on cardiac gene expression. At 24 hpf and at 33 hpf heart tube, no alternation was detected in cmlc2 (A, B, E, F), tbx5 (C, D) or nppa (G, H) expression level between wild-type (A, C, E, G) and ndrg4 morphants (B, D, F, H) with whole mount in situ hybridization. Note normal jogging out in the heart of morphants. At 48 hpf heart, no change was observed in vmhc (I, J), cmlc2 (M, N), amhc (Q, R) and nppa (K, L) expression level between wild-type (I, K, M, O, Q, S) and ndrg4 morphant (J, L, N, P, R, T). nppa and cmlc2 are expressed throughout the heart, vmhc is expressed in the ventricle and amhc is expressed in the atrium. Note the unlooped heart of ndrg4 morphant (N). At 48 hpf, versican is expressed at the boundary of the atrium and the ventricle (arrow) in wild type embryos (O), but is expressed throughout ndrg4 morphrant hearts (P). bmp4 expression is also prominent in the myocardial region of AV junction (between the arrows) in 48 hpf wild type zebrafish embryos (S), whereas ndrg4-MO2 injection resulted in strong ectopic expression of bmp4, especially in the ventricular region (T, arrowhead). (A, B) Dorsal views with head towards the top; (C–H), Lateral views with anterior towards the left; (I–T), frontal views with anterior towards the top. |
|
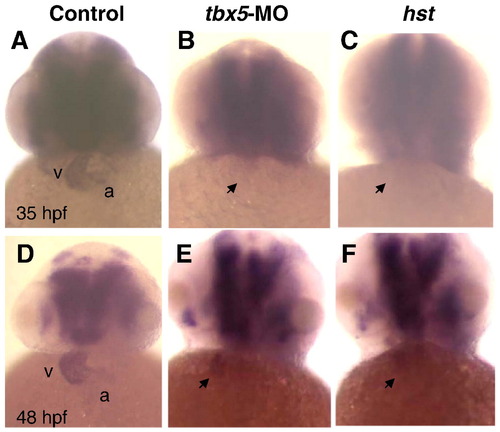
ndrg4 is down regulated by Tbx5 attenuation. At 35 hpf (A–C) and 48 hpf (D–F), compared with uninjected (A, D) embryos, ndrg4 expression (whole mount in situ hybridization) in tbx5-MO injected (B, E) or hst mutant (C, F) embryos was dramatically decreased. Arrow indicates heart region. a, atrium; v, ventricle. All are frontal views with anterior towards the top. |
|
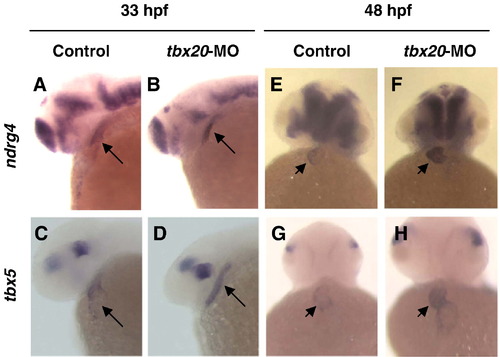
ndrg4 is up regulated by Tbx5 augmentation via tbx20-MO. Compared to uninjected embryos, tbx5 and ndrg4 expression in tbx20-MO injected embryos at 33 hpf (A–D; lateral views with anterior towards the left) and 48 hpf (E–H; frontal views with anterior towards the top) was significantly increased. Arrow indicates heart and arrowhead indicates ventricle of heart. |
|
In vivo validation of NDRG4 MO1 (knockdown by translation-blocking). (A) Schematic of the strategy to target the translational start site with NDRG4 MO1. (B) Schematic map of a transgenic construct in which the cmlc2 promoter is upstream of an NDRG4-EYFP fusion DNA. H19 insulators in the pWhere plasmid (InvivoGen) flanked the cmlc2-NDRG4-EYFP DNA fragment, which includes the binding region of NDRG4,/em> MO1. (C) After Co-injection of the linearized DNA with 6.4 ng of control MO (a) or NDRG4 MO1 (b) into wildtype embryo at one-cell stage, green fluorescent signals due to NDRG4-EYFP expression in cardiomyocytes were observed in 7.9% (control MO) or 0% (NDRG4 MO1) of injected embryos at 48 hpf. Arrow indicates the heart region. |

Unillustrated author statements EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 317(2), Qu, X., Jia, H., Garrity, D.M., Tompkins, K., Batts, L., Appel, B., Zhong, T.P., and Baldwin, H.S., ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish, 486-496, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.