- Title
-
Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest
- Authors
- Minchin, J.E., and Hughes, S.M.
- Source
- Full text @ Dev. Biol.
|
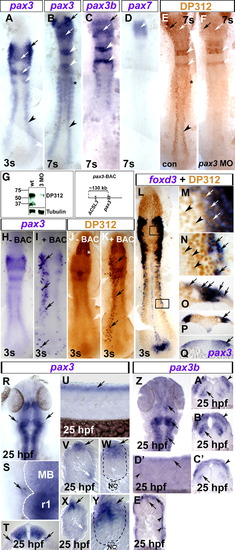
Pax3/7 expression in neural crest. In situ hybridisation for pax3 (A, B, H, I, Q–Y), pax3b (C, Z–E′), pax7 (D), foxd3 (L–P) and immunodetection of Pax3/7 by DP312 (E, F, J–P brown). Dorsal flat mounts (A–F, H–N, R, S, Z) have anterior to top. Lateral flat mount (U, D′) have dorsal up and anterior to left. Transverse cryosections (O–Q, T, V–Y, A′–C′, E′) have dorsal up. (A) At 3 s, pax3 is expressed in delaminated cranial NC (black arrow), lateral neural plate (black arrowhead) and the dorsal CNS, corresponding to posterior diencephalon/midbrain and rhombomeres 2/4 (white arrows; Seo et al., 1998). (B–D) At 7 s, pax3 mRNA is still detected in cranial NC (black arrows), neural plate (black arrowhead), rhombomeres (white arrowheads), dorsal brain regions (white arrows) and somites (black asterisk). Pax3b and pax3 mRNAs coincide in cranial NC (black arrow), rhombomeres 2/4 (white arrows) and dorsal brain (white arrows), but not in trunk NC. Pax7 mRNA is only expressed in the midbrain at 7 s (D, white arrow). (E, F) Pax3/7 immunoreactivity (DP312) corresponds to regions of pax3, pax3b and pax7 expression in wild type (E, compare to panels B–D). Pax3 MO injection ablated DP312 immunoreaction where pax3 mRNA does not overlap with expression of other pax3/7 genes (F, compare to black arrowhead in panel B), and reduced immunoreaction in cranial regions (arrows) and rhombomeres 2/4 (white arrowheads). (G) Western blot showing reduction in DP312 immunoreactivity in pax3 MO-injected embryos at 24 hpf, whereas tubulin loading control is unaltered. Zebrafish Pax3 and Pax7 have predicted Mr 55.7 kDa, Pax3b 53.3 kDa and Pax7b 56.3 kDa. The band at 37 kDa could represent alternate pax3/7 isoforms or degradation products. Note that the control lane is reprinted from Hammond et al. (2007). (H–K) Injection of a BAC containing pax3 (pax3-BAC, map above right) to wild type embryos results in mosaic over-expression of pax3 mRNA (I, black arrows) and protein (K, black arrows) in endogenous pax3 locations, demonstrating that DP312 recognises zebrafish Pax3. Note the lack of extra immunoreactivity in eye fields, in which DP312 reacts with Rx proteins (white asterisk; [Davis et al., 2005] and [Pujic and Malicki, 2001]). (L–Q) Foxd3 mRNA, a NC marker, co-localises around nuclear DP312 immunoreactivity (L–P). High magnification images of DP312 from black boxes in panel L in cranial NC cells (M, white arrows) and also neural crest precursors in the trunk lateral neural plate (N, black arrows). Pax3/7+ cells lacking foxd3 mRNA are also present in dorsal neural plate in cranial and trunk regions (M, N, respectively; black arrowheads). Transverse cryosections reveal that foxd3 mRNA (O, P, arrows) is located in the lateralmost region of highest pax3 mRNA (Q, arrow) accumulation in the neural plate, and in underlying delaminated NC cells. (R–T) Expression of pax3 outside the neural tube in putative ophthalmic lobe of trigeminal ganglion at 25 hpf (R–T; black arrows; white dashed line shows CNS boundary in panel S; MB midbrian, r1 rhombomere 1). (U–Y) In trunk (V, W) and tail (X, Y) regions, pax3 is expressed in the dorsal neural tube (black arrows) and lateral somite (white arrows). Note the lack of strong punctate pax3 expression in migrating NC cells. Black dashed lines show neural tube boundary. NC = notochord. (Z–E′) Pax3b mRNA is detected in dorsal neural tissue in head (Z–C′: arrows, neural laminae; arrowheads, NC) and trunk (D′, E′: arrows, neural tube; arrowheads, superficial somite region). EXPRESSION / LABELING:
|
|
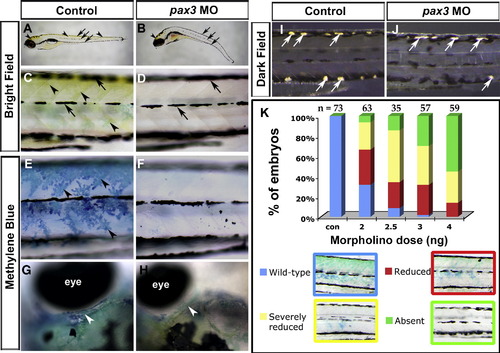
Pax3 knockdown blocks terminal differentiation of trunk xanthophores whilst other pigment cell types differentiate. Live control (A, C, E, G, I) or pax3 MO-injected (B, D, F, H, J) zebrafish larvae showing yellow xanthophore pigmentation in brightfield images (A–D), stained with methylene blue to reveal pterinosomes (E–H) or in darkfield to reveal iridophores (I, J). Lateral views (A–F, I, J) are anterior to left and dorsal to top. Dorsal views of head region (G, H) are anterior to left and lateral to top. (A–D) At 5 dpf, yellow xanthophore pigmentation is visible on the dorsal head and trunk (arrowheads). Embryonic melanophores are arranged in stripes (arrows). In pax3 morphants, no yellow xanthophore pigmentation is visible in the trunk and is reduced in the dorsal head (B, arrowhead), whereas embryonic melanophore stripes are still present and located correctly (B, D, arrows). (E–H) At 5 dpf, blue pterinosomes (arrowheads) coincide with xanthophore locations in dorsal and posterior regions overlying epaxial and hypaxial somite in the trunk (E) and above the eye in the head (G, white arrowhead). In pax3 morphants, pterinosome stain is absent from the trunk (F) and reduced in the head (H, arrowhead). (I, J) At 6 dpf, shiny iridophore pigment cells (white arrows) have begun to differentiate and are located at the dorsal and ventral larval melanophore stripes (I). Overlying xanthophores give the iridophores a yellow tint. In pax3 morphants, iridophores differentiate in both dorsal and ventral positions but appear white due to the lack of overlying xanthophores (J). (K) Severity of methylene blue-stained xanthophore phenotype in trunk is dependent on dose of pax3 MO. Overall reduction in trunk xanthophores (reduced) was distinguished from complete absence in most of the trunk (severely reduced). Number of embryos analysed (n) are given above columns. |
|
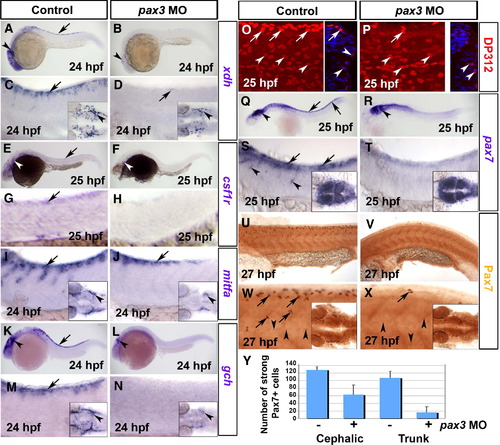
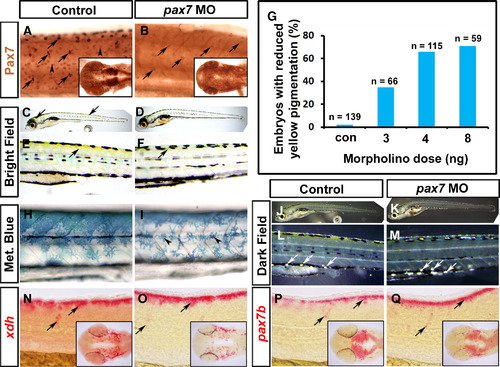
Pax3 morphants have a defect at early stages of xanthophore development. Control (A, C, E, G, I, K, M, O, Q, S, U, W) and pax3 MO-injected (B, D, F, H, J, L, N, P, R, T, V, X) 1-day-old zebrafish embryos analysed by in situ mRNA hybridisation for xdh (A–D), csf1r (E–H), mitfa (I, J), gch (K–N) or pax7 (Q–T) or by immunodetection for Pax3/7 with DP312 (O, P) or anti-Pax7 (U–Y). Lateral views anterior to left. Insets show dorsal views of the head region. (A–D) Pax3 MO essentially ablates xdh mRNA in trunk (arrows), but reduced expression persists in the head (arrowheads). (E–H) Csf1r mRNA is undetectable in pax3 morphant trunk. Although residual expression is present in pax3 morphant head, it appears differently localised compared to xdh (white arrowheads). (I, J) In control embryos, expression of the chromatophore marker mitfa shows the start of NC migration in anterior trunk. In pax3 morphants, mitfa expression is severely reduced, but less so than xdh and csf1r. (K–N) Gch mRNA is strongly depleted in trunk of pax3 morphants (arrows), but is still detectable in head (arrowhead). (O, P) At 25 hpf, DP312 detects cells in the dermomyotome weakly (arrowheads) and pre-migratory NC cells intensely (arrows). In pax3 morphants, the strongly-labelled NC cells are dramatically reduced, whereas dermomyotome signal remains. Note the co-localisation of Pax3/7 and Hoechst 33258 DNA stain in transverse cryosections of one side of the trunk at right. (Q–T) At 25 hpf, pax7 mRNA is lost from trunk NC cells, but retained in dermomyotome and neural tube. (U–X) Immunodetection of Pax7 at 27 hpf demonstrates that intensely labelled cells migrate in an anterior to posterior wave on the lateral surface of the somite, resembling NC. Pax7+ cells are severely reduced or absent in the trunk of pax3 morphants, and mildly affected in head regions (insets). (Y) Counts of strongly Pax7+ NC cells quantify the difference in severity between head and trunk regions. |
|
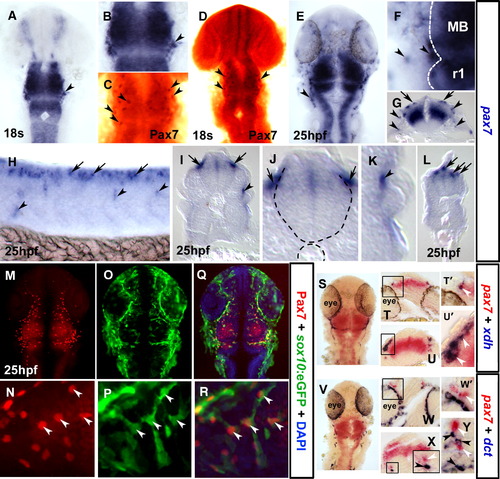
Pax7 is a marker of the xanthophore lineage. In situ mRNA hybridisation for pax7 (A, B, E–L, S–Y), xdh (S–U′) and dct (V–Y) and immunodetection of Pax7 (C, D, M, N, Q, R) and sox10:GFP (O–R). Dorsal flat mounts (A–F, M–S, V) are anterior to top. Lateral flat mount (H) is anterior to left and dorsal to top. Transverse cryosections (G, I–L, T, U, W–Y) are dorsal to top. (A–D) Pax7 mRNA is detected at 18 s in NC-like cells at the mid-hindbrain boundary (A, B; arrowheads), concomitant with Pax7 protein in cranial NC-like cells (C, D; brown, arrowheads). (E–L) By 25 hpf, pax7 mRNA is detected in pre-migratory (arrows) and laterally migrating (arrowheads) NC-like cells in head (E–G), trunk (H, shown in section I and magnified J, K) and tail (L) regions. (M–R) Co-localisation of GFP from the sox10:egfp transgenic line with Pax7 protein demonstrates that Pax7 marks NC cells (arrowheads). (S–Y) Co-localisation in NC of pax7 (white arrowheads) and xdh but not dct (black arrowheads) mRNAs demonstrates that pax7 is expressed in the xanthophore lineage. Cryosections at the level of the forebrain (T, W) or hindbrain (U, X, Y), with the boxes in panels T, U, W and panel X magnified in panels T′, U′, W′ and panel X inset, respectively. EXPRESSION / LABELING:
|
|
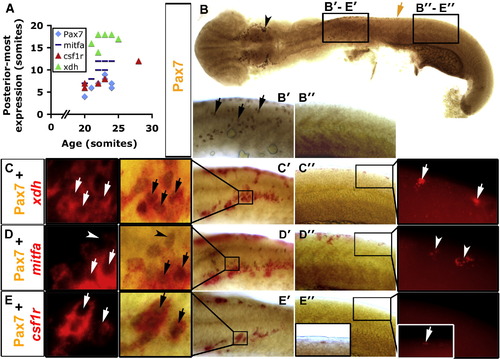
Xanthophore specification occurs before accumulation of Pax7. Immunodetection of Pax7 (brown, B–E) or in situ mRNA hybridisation (fast red fluorescence) for xdh (C), mitfa (D) or csf1r (E). Whole 20 s embryo flatmounts with anterior to left show dorsal and lateral views in head and trunk regions, respectively. Panels B′–E′ are anterior trunk, panels B″–E″ are posterior trunk according to position of black boxes in panel B. (A) Graph showing posteriormost extent of each marker in relation to embryonic stage. (B) Pax7+ NC are detected in the head (arrowhead) and in pre-migratory positions in the anterior trunk (black arrows in panel B′), but not in more posterior regions (B″). Brown arrow indicates the posteriormost extent of Pax7+ NC. (C–E) Dual stain for Pax7 and indicated mRNA showing anterior (C′, D′, E′) and posterior (C″, D″, E″) trunk in brightfield. The insets (C′–E′, C″–E″) are magnified to reveal fast red and its fluorescence. (C) xdh is expressed together with Pax7 (arrows) in anterior trunk, but alone in posterior trunk (C″ inset, white arrows). (D) mitfa is expressed in anterior and posterior trunk. Some Pax7+ NC co-express mitfa in anterior trunk, others do not (D′, arrows and arrowheads, respectively). mitfa is also expressed alone, probably in melanoblasts (Lister et al., 1999; D″, arrowheads). Mitfa expression is lateral to the Pax7+ cells in anterior trunk NC at this stage (D′). (E) csf1r mRNA is detected only in Pax7+ NC in anterior trunk (E′, arrows). However, the posteriormost Pax7+ NC are csf1r+ (E″, inset, white arrow). EXPRESSION / LABELING:
|
|
Xanthophore pigmentation is reduced after knockdown of Pax7. Immunodetection of Pax7 (A, B), live images (C–F, H–M) and in situ mRNA hybridisation for xdh (N, O), pax7b (P, Q) of larvae. Lateral flatmounts are anterior to left and dorsal to top (A–F, H–Q) and dorsal flatmounts of head are anterior to left (insets, A, B, N–Q). (A, B) Pax7 protein is detected in migrating NC (arrows) and dermomyotome cells (arrowheads) at 26 hpf (A). Injection of a morpholino specific to pax7 results in ablation of Pax7 in dermomyotome cells and severely reduced reactivity in brain (inset) and NC (B, arrows). (C–F) At 6 dpf, yellow xanthophore pigmentation is present in the dorsal head and trunk (C, E; arrows). Pax7 morphants, show a marked reduction in yellow pigmentation in both head and trunk regions (D, F). (G) Dose-dependency of reduction in yellow pigmentation in pax7 morphants. (H, I) Methylene blue reveals morphological defects in pterinosome-containing cells after pax7 MO injection (I, arrowheads). (J–M) Dark field images at 6 dpf reveal iridophores in both control and pax7 morphants (arrows). (N–Q) xdh (N, O) and pax7b (P, Q) mRNAs persist in pax7 morphant xanthophore precursors (arrows). |
|
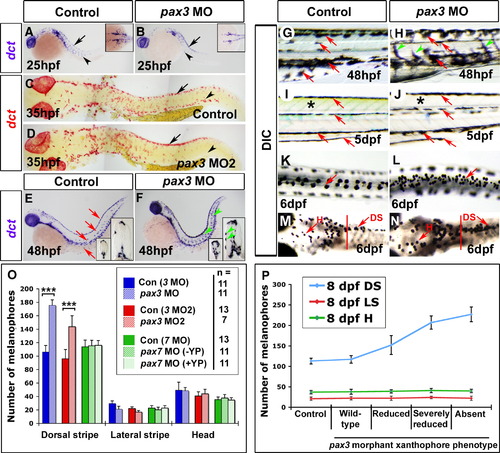
Delayed melanophore origin then increase correlate with xanthophore loss in pax3 morphants. Embryos analysed by in situ mRNA hybridisation for dct (A–F), live (G–J) or fixed (K–N) shown with anterior to left. Wholemounts are shown in lateral (A, B, E–J; dorsal to top) or dorsal (K–N) view. Whole embryo flatmounts (C, D) show dorsal and lateral views in head and trunk regions, respectively. (A, B) At 25 hpf, dct mRNA is detected in pre-migratory (arrows) and migrating melanoblasts (arrowheads) in the head and trunk, and also in the eye pigment epithelium. In pax3 morphants, dct+ melanoblasts are reduced, but still able to migrate (B, arrowhead). (C, D) At 35 hpf, substantially more dct+ melanoblasts are situated dorsally in pax3 MO2-injected embryos (arrows), but migrated dct+ melanoblasts are reduced (arrowheads). (E–H) At 48 hpf, dct mRNA is present in dorsal, lateral and ventral stripes, and in single cells on yolk sac, matching the melanophore pattern (E, G red arrows). In pax3 morphants, melanised and dct+ cells are more abundant between stripes on the medial and lateral pathways (F, H arrowheads). (I, J) At 5 dpf in control embryos, four larval melanophore stripes are clearly defined (I, red arrows) and the trunk has a yellow/green cast due to xanthophore pigmentation (I, asterisk). In pax3 morphants, the trunk is devoid of xanthophore pigmentation (J, asterisk), but the melanophore pattern is within normal variation (J, red arrows). (K-N) Treatment of 6 dpf embryos with adrenalin causes melanophore contraction that permits cell counting. In trunk (K, L) but not head (M, N anterior to red bar), pax3 MO-injection increases melanophore number in the dorsal stripe. (O) Quantification of melanophore number in pax3 MO, pax3 MO2 and pax7 MO-injected embryos at 6 dpf. pax7 morphants were categorised into normal yellow pigmentation (+ YP) or reduced yellow pigmentation (- YP) groups. ***p < 0.0001. (P) Correlation of melanophore increase in pax3 morphants with loss of xanthophores. Pax3 MO2-injected 8 dpf larvae were categorised by severity of methylene blue-stained xanthophore phenotype, and melanophores counted. DS = dorsal stripe, LS = lateral stripe, H = head melanophores. |
|
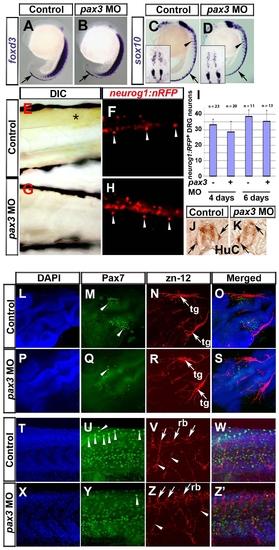
Pax3 is required for enteric nervous system development. In situ mRNA hybridisation analysis of foxd3 (A–D, G, H) and sox10 (E, F, I–L) and immunodetection of DP312 (A–F) and HuC (M–P). Dorsal flat mounts (A–F, I, J upper insets) are anterior to top. Lateral view whole mounts are anterior to top and dorsal to right (G–J) or anterior to left dorsal to top (K, L). Transverse cryosections (G–J lower insets, M–P) are dorsal to top. (A–F) Injection of pax3 MO depletes nuclear Pax3/7 immunoreaction at 3 s (B, D, F; arrows) in regions of pax3 mRNA expression, but leaves residual signal in regions of expression of pax3b, pax7 or Rx genes in the brain and eye (asterisks). Foxd3 expression in NC, tailbud and floorplate is unaffected by Pax3 knockdown (A, B; cranial NC, boxes enlarged in panels C, D). Pax3 knockdown has no effect on sox10 expression in cranial NC and otic placode at this stage (E, F). (G–J) At 25 hpf, foxd3 expression is restricted in NC to a cluster of nascent pre-migratory cells in the tail (G, H, arrows) and to lateral line primordium (G, H, arrowheads). Pax3 MO injection has no effect on foxd3 expression in either location (H), but reduces sox10 mRNA in pre–migratory crest and cells migrating on the medial pathway (I, J, white arrows), while having little effect on expression in the head (I, J, black arrows). Significant sox10 mRNA in nascent crest (I, J, white arrowheads) is down-regulated anteriorly. (K, L) At 36 hpf, sox10 mRNA is reduced in pax3 morphants in the ventral head region destined to form enteric neurons (arrows) but is still detected in otic placode and glia (arrowheads). (M–P) Pax3 MO injection depletes enteric neurons (arrows) revealed by immunodetection in anterior (M, O) or posterior (N, P) gut. EXPRESSION / LABELING:
PHENOTYPE:
|
|
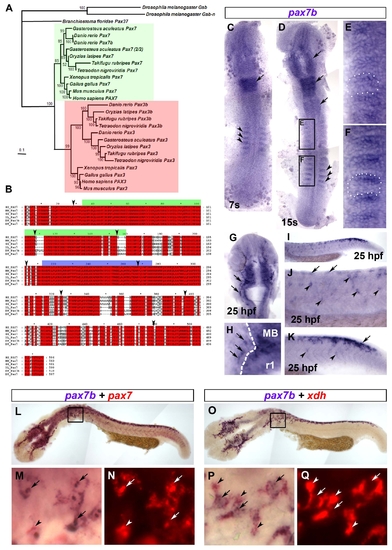
Phylogenetic analysis and expression of pax7b. A. Bayesian phylogenetic analysis of the Pax3/7 sub-family. The tree was rooted with D. melanogaster Gsb and Gsb-n. Confidence for nodes is indicated by Bayesian support values. Green and red boxes indicate Pax7 and Pax3 sub-groups, respectively. B. Multiple sequence alignment of Pax7b sequence with homologous Pax7 proteins. The green and blue bars above the sequences indicate the Paired box and homeodomain, respectively. Arrowheads denote exon boundaries. Red boxes denote identical residues. Grey boxes show similar residues. HS = Homo sapiens, MM = Mus musculus, GG = Gallus gallus, XL = Xenopus leavis, DR = Danio rerio. C-Q. In situ mRNA hybridisation analysis of pax7b (C-Q) alone or co-localised with either pax7 (L-N) or xdh (O-Q), Dorsal view flatmounts are anterior to top (C-H) and lateral view flatmounts are anterior to left and dorsal to top (I-K,L-Q). At 7s, pax7b is expressed in anterior rhombomeres (C,D arrow) and the anterior edge of the newly-formed somites (C,D, arrowheads). By 15s, pax7b expression is maintained in rhombomeric stripes and appears in midbrain and anterior spinal cord (D, arrows). Expression in somites (black boxes, magnified in E,F, white dots delineate a somite) is reduced in older somites (E), but abundant in the anterior region of young somites (F). At 25 hpf, pax7b is expressed in cranial NC (G,H arrows; MB = midbrain, r1 = rhombomere 1; white dashes CNS border). In trunk NC, pax7b is expressed primarily in nascent pre-migratory cells (arrows) in tail (I enlarged in K) and older migratory cells (arrowheads) in trunk (I enlarged in J). Pax7b co-localises with both pax7 (L-N) and xdh (O-Q) mRNA. Boxes (L,O) are magnified in M,N,P,Q to show regions of overlapping (arrows), and non-overlapping (arrowheads) mRNA. Xdh mRNA is mainly cytoplasmic (arrowheads, P,Q), whereas pax7b and pax7 mRNAs show prominent nuclear puncta (arrows, M,P). EXPRESSION / LABELING:
|
|
Pax3 is not required for sensory neural crest lineages. In situ mRNA hybridisation analysis of foxd3 (A,B) and sox10 (C,D), live bright field (E,G) and fluorescent (F,H) analysis of the neurog1:nRFP transgenic fish line and immunodetection of HuC (J,K), Pax7 (M,Q,U,Y) and zn-12 (N,R,V,Z). Lateral view wholemounts are anterior to top and dorsal to right (A-D). Lateral view flatmounts are anterior to left and dorsal to top (E-H, T-Z′). Dorsal view flatmounts are anterior to top (C,D insets) or lower left (L-S). Transverse cryosections are dorsal to top (J,K). A-D. At 18s, foxd3 mRNA is detected in pre-migratory NC (arrows), whereas sox10 mRNA is in both pre-migratory and migrating NC (arrowheads). Expression of foxd3 and sox10 appears unaffected in pax3 morphants (B,D). E-K. In 6 dpf larvae, xanthophore pigmentation is present in the trunk of neurog1:nRFP transgenic fish (E, asterisk), in which DRG neurons (F, arrowheads) and weaker CNS neurons are visible. Injection of pax3 MO ablates xanthophore pigmentation (G) but DRG neurons remain (H, arrowheads). Quantification of DRG neuron number reveals that there is no significant difference between control and pax3 morphants (I). DRG neurons are sometimes mis-positioned dorsally in pax3 morphants (J,K arrows). L-Z′. At 24 hpf, Pax7+ NC cells are clustered in the head of control embryos (M, arrowheads) near to zn-12 immunoreactive trigeminal ganglia (N, arrows, tg). In pax3 morphant, there are fewer Pax7+ NC cells in cranial regions (Q, arrowhead) but the trigeminal ganglia have formed normally (R, arrows). In trunk regions, Pax7+ NC cells are situated in the pre-migratory position (U, arrowheads). Zn-12 staining shows the Rohon-Beard neuronal cell bodies (V, arrows, rb) and processes (V, arrowheads). Pax3 morphants lack Pax7+ pre-migratory NC cells (Y), whereas Rohon-Beard neuronal cell bodies and processes (Z) and dermomyotomal Pax7 accumulation (Y) appear normal. EXPRESSION / LABELING:
|
|
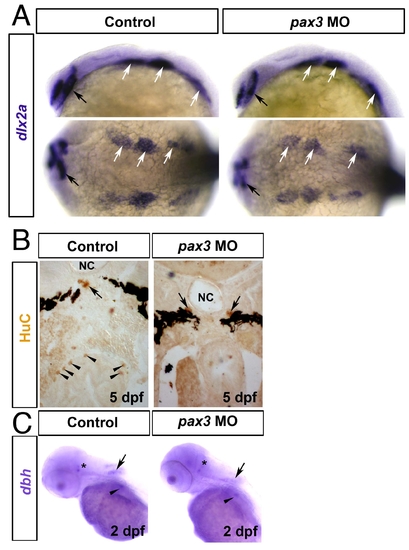
Ectomesenchymal and sympathetic neural crest is unaffected in pax3 morphants. In situ mRNA hybridisation for dlx2a (A), dbh (C) and immunodetection of HuC (B). Wholemounts in lateral (A upper panels,C; anterior to left and dorsal to top) or dorsal (A lower panels; anterior to left) view. Transverse sections of the foregut at hindbrain level are dorsal to top (B). A. At 24 hpf, dlx2a mRNA is detected in forebrain (black arrows) and branchial arch ectomesenchymal NC (white arrows) and is unaffected by pax3 MO injection. B. Bilateral sympathetic neurons (arrows) located between the notochord (NC) and ventral melanophore stripes are unaffected in a 5 dpf pax3 morphant lacking enteric neurons (arrowheads) around the gut. C. At 48 hpf in control embryos, dbh mRNA is present in cranial sympathetic neurons (asterisk), neurons of the dorsally located medulla oblongata (arrow) and the cervical sympathetic ganglia (arrowhead). In pax3 morphants, dbh mRNA is unaffected. |

Unillustrated author statements |
Reprinted from Developmental Biology, 317(2), Minchin, J.E., and Hughes, S.M., Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest, 508-522, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.