- Title
-
Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis
- Authors
- Reifers, F., Bohli, H., Walsh, E.C., Crossley, P.H., Stainier, D.Y., and Brand, M.
- Source
- Full text @ Development
|
Expression of Fgf8 in wild-type embryos. (A) Fgf8 in the blastoderm margin at 30% epiboly. (B,C) At shield stage, Fgf8 is expressed in a dorsoventral gradient in the germ ring with a high point of expression in the shield (B, vegetal pole view; C, lateral view). (D,E) Graded expression persists in the margin of the blastoderm at 90% epiboly. Prospective anterior hindbrain expresses Fgf8 (D, dorsal view; slightly tilted in E). (F) Forebrain expression in a tailbud stage embryo (arrowheads point to high points of expression). (G) Prospective MHB domains fuse at the midline at tailbud stage. (H) 4-somite stage, lateral view. Expression in forebrain, mid-hindbrain region, segmental plate and tailbud. (I,J) Flat mount of H, depicting anterior and posterior expression domains. (K) Fgf8 expression at the anterior somite border (arrowheads). (L) Flat-mounted 19-somite embryo. Expression in the heart ring posterior to the MHB. (M) Flat mount at 22 somites; expression in the brain is detected at the MHB, dorsal diencephalon, retina and optic stalks. (N) Lateral view of a 24 hours embryo. Additional expression occurs in the facial ectoderm. (O) Thick cross section through the MHB demonstrating absence of expression in floorplate. (P) Lateral view of a dissected brain at 36 hours of development. Additional expression in the infundibulum, hypophysis and otic vesicle (eyes are removed). (Q) Details of expression in the retina, choroid fissure and the optic stalks. Additional expression is detected in nasal ectoderm and the hyoid. (R) Ventral view of head at 36 hours demonstrating expression in the retinal epithelium, but not in the lens. (S) Frontal view, expression in the facial and nasal ectoderm. cf, choroid fissure; dd, dorsal diencephalon; fb, forebrain; fec, facial ectoderm; fp, floorplate; ht, heart; hy, hyoid; hyp, hypophysis; inf, infundibulum; ls, lens; mb, midbrain; mhb, mid-hindbrain boundary; nec, nasal ectoderm; os, optic stalks; ov, otic vesicle; ret, retina; sh, shield; som, somites; tb, tailbud, yp, yolk plug. EXPRESSION / LABELING:
|
|
Fgf8 expression in early midbrain and anterior hindbrain development. (A-D) Double ISH with Fgf8 (blue) and Krox20 (red, fluorescent) of wild-type embryo at tailbud stage (A,B) and 5 somites (C,D). At tailbud stage, Fgf8 expression extends throughout the anterior hindbrain incl. rhombomere 4 (r4) posteriorly. At 5 somites, expression of Fgf8 is detected at the MHB, in r1, r4 and in ventral r2 (see also Fig. 2H). (E-J) Double ISH with Fgf8 (blue) and Pax2.1 (red, fluorescent) at 90% epiboly (E-H) and 6 somites (I,J). At 90% epiboly, the Fgf8 expression domain is located posterior to the Pax2.1 domain with very little overlap (E,F; higher magnification: G,H), while at 6 somites the Fgf8 domain at the MHB is completely included in the Pax2.1 expression domain (visible as quenching of the fluorescent Pax2.1 signal). Embryos in C-J are flat mounted, A,C,E,G and I show bright field, B,D,F,H and J show fluorescent images of the same embryos. EXPRESSION / LABELING:
|

RT-PCR analysis of Fgf8 transcripts in wild-type and acerebellar embryos. (A) Structure of wild-type cDNA and placement of primers. (B) Only transcripts containing exon 2 are detected in wild type throughout development either with primers spanning exon 2 (P5 + P6) or with one primer located in exon 2 (P7 + P6). (C) Exon 2 is missing in Fgf8 transcripts of acerebellar embryos. RT-PCR with primers spanning exon 2 (P5 + P6) yields a single band of 612 bp in wild-type embryos. In acerebellar embryos, no transcripts of wild-type size are detected; instead only transcripts without exon 2 are seen, which are shorter by 107 bases (compare lanes 2 and 4, and lane 5 and 6). In heterozygous siblings, bands of both sizes are detectable (lane 3). With one primer located in exon 2 (P5 + P8), amplification is only possible in acerebellar embryos after reamplification (compare lane 9 and 12), while in wild-type embryos and siblings one round of amplification is sufficient (lanes 7, 8), demonstrating the low abundance of exon 2 containing transcripts in acerebellar embryos. (D) acerebellar mutant embryos and their siblings were sorted by phenotype, indicated by bars. Single embryo RT-PCR with primers spanning exon 2 (P5 + P6) was performed for 101 acerebellar embryos and 13 siblings. Phenotypically wild-type siblings show either a wild-type or a heterozygous pattern. In all phenotypically acerebellar embryos tested, only exon 2-less transcript is detected, confirming genetic linkage (0 ± 0.5 cM). |
|
Function of wild-type and mutant Fgf8 in dorsoventral patterning of the gastrula. Adaxial and somitic mesoderm is visualized with myoD (blue), or intermediate mesoderm and MHB with Pax2.1, and location of the lacZ co-injected cells with an antibody to β-gal (brown). RNA distribution is mostly restricted to one side, allowing comparison with the contralateral side as a control. (A-F) Misexpression of Fgf8 in wild-type embryos by RNA injection. (A,B) Live 28 h embryo with axis duplication (A) and stained for myoD (B). (C) Axial location of the injected RNA yields no obvious defect in mesoderm patterning. (D) Expansion of somitic mesoderm on the injected side of embryo. (E) Pax2.1-positive intermediate mesoderm (black arrowheads) is missing on the injected side (white arrowheads). (F) Fgf8 misexpression alters the d/v, but not the a/p extent of the Pax2.1 expression domain on the left, injected side (midline is given by a dashed line; op, otic placode). (G) No effect is seen after injection of the acerebellar version (lacking exon2) of Fgf8 RNA, showing that the acerebellar transcript is inactive in vivo. (H) lacZ control injections had no effect. (I) Summary of the effects observed after injection of Fgf8. Left: schematic fate map of a gastrula, and the Fgf8 gradient in the germ ring. Right: consequence of misexpressing Fgf8 in the respective area. All embryos shown are at early to midsomitogenesis stages; BD, G, H show myoD in situ stainings of injected embryos, E and F show Pax2.1 in situ stainings; β-gal was detected by antibody staining. |
|
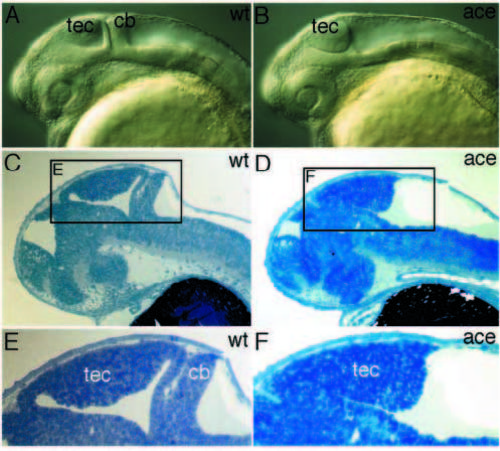
Brain phenotype of acerebellar embryos. (A,B) At pharyngula stage, mutant embryos lack a cerebellum and the midhindbrain fold, but show an enlarged tectum (lateral view of living embryos). (C,D) Sagittal section of 36 hour embryos. (E,F) High magnification view of area depicted in C and D, showing the midhindbrain phenotype in more detail. cb, cerebellum; tec, tectum. PHENOTYPE:
|
|
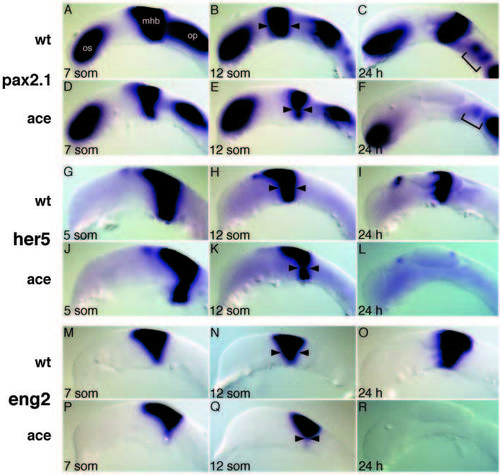
Fgf8 is required for the maintenance of MHB marker genes. Lateral views of dissected brain primordia. Stages and markers as indicated. (A-F) Expression of Pax2.1 in wild-type (A-C) and acerebellar (D-F) embryos. Notice the gradual reduction in width at the MHB in B, E, and the reduction of otic placode (A,D), optic stalk and anterior hindbrain expression in C versus F. (G-L) Expression of Her5 in wild-type (G-I) and acerebellar (J-L) embryos. Expression of Eng2 in wild-type (M-O) and acerebellar (P-R) embryos. Arrowheads depict the width of the MHB, brackets mark anterior hindbrain. op, otic placode. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fgf8 expression at the MHB is independent of Pax2.1. (A) Expression of Fgf8 at the MHB in noi/Pax2.1 mutant embryos cannot be distinguished from their wild-type siblings at the 10- somite stage. (B,C) At 18 somites, the expression of Fgf8 is absent from the MHB in noi/Pax2.1 mutant embryos, due to the loss of MHB territory. A-C show lateral views of Fgf8 in situ-stained embryos. ht, heart; mhb, mid-hindbrain boundary. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fgf8 is involved in mesoderm and somite patterning. (A,B) Expression of myoD is strongly reduced in adaxial mesoderm of acerebellar embryos at 80% epiboly (arrowheads point to remnants of expression). (C,D) At tailbud stage, myoD expression in adaxial mesoderm is interrupted in acerebellar (arrowheads). (E,F) myoD staining in the somitic mesoderm is strongly reduced in mutants at the 10- somite stage. (G,H) At 24 hours, the expression of myoD is weak in the smaller and less-well-differentiated somites of acerebellar embryos. (I,J) No obvious difference could be detected between wild-type and acerebellar embryos in formation of intermediate mesoderm, shown here with Pax2.1 staining at the 7-somite stage. (K,L) Expression of snail1 is reduced in acerebellar embryos in the region around the tailbud at 6- somite stage (arrowheads point to the wild-type border of expression). (M,N) At the 13-somite stage and (O,P) 20-somite stage, snail1 transcripts are strongly reduced in the somites of mutant embryos. (Q,R) Dorsal view of wild-type embryo stained for eng2 (blue) and Fgf8 (red, fluorescent) showing partial overlap of these expression domains at an early stage of somite development (arrows). Note the restriction of Fgf8 expression to anterolateral cells of the somites over time (anterior is to the left; brackets depict adaxial cells; nc, notochord). (S,T) Muscle pioneers are reduced in acerebellar embryos, as shown here for 24 hours embryos with Eng2 staining. EXPRESSION / LABELING:
PHENOTYPE:
|
|
acerebellar embryos show no severe defects in pectoral fin development. (A-D,M) Fgf8 expression in the finbud. (A-C) Wild type. (D) acerebellar. No Fgf8 expression is detected at 30 hours of development. (E-G) shh expression in the ZPA precedes Fgf8 expression and is not affected in acerebellar. (E,F) Wild type. (G) acerebellar. (H-J) eng1 (arrowheads) in the ventral fin bud precedes Fgf8 and is normal in acerebellar. (H,I) Wild type. (J) acerebellar. (K,L) Fins of wild-type (K) and acerebellar (L) embryos on day 5 of development are of similar size and shape. (M) Fgf8 expression in the distalmost ridge, AER, of the developing fin at 48 hours of development (viewed from posterior). a, anterior; dis, distal; dor, dorsal; p, posterior; pro, proximal; ven, ventral. |