- Title
-
Anteroposterior patterning in the zebrafish, Danio rerio: an explant assay reveals inductive and suppressive cell interactions
- Authors
- Sagerström, C.G., Grinblat, Y., and Sive, H.
- Source
- Full text @ Development
|
Marker gene expression in cultured animal caps. (A) Schematic representation of animal cap dissection and culture. Animal caps were dissected from sphere stage embryos (late blastula, 4 hpf), aggregated into groups of 10 and harvested immediately after dissection or when control embryos reached the onset of gastrulation (6 hpf, 2 hours in culture), the end of gastrulation (10 hpf, 6 hours in culture) or the end of somitogenesis (24 hpf, 20 hours in culture). (B) RTPCR analysis of gene expression in explanted animal caps and whole embryos. Explants (lanes 1- 4) and whole embryos (lanes 5-8) were harvested at the time of dissection (lanes 1 and 5), or after 2 hours (lane 2 and 6), 6 hours (lanes 3 and 7) or 20 hours in culture (lane 4 and 8) and analyzed for expression of the mesodermal markers ntl (Schulte-Merker et al., 1992), gsc (Stachel et al., 1993; Schulte-Merker et al., 1994a; Thisse et al., 1994), eve (Joly et al., 1993) and MyoD (Weinberg et al., 1996), the anterior marker otx2 (Li et al., 1994), the neural markers zash1a (Allende and Weinberg, 1994), zash1b (Allende and Weinberg, 1994), pax6 (Krauss et al., 1991; Puschel et al., 1992), wnt1 (Molven et al., 1991), eng3 (Ekker et al., 1992), krx20 (Oxtoby and Jowett, 1993), the type I cytokeratin cyt1 (Sagerström and Sive, unpublished data) and the ventral marker gta3 (Neave et al., 1995) by relative quantitative RT-PCR as described in Materials and Methods. Relative quantitation enables comparison of the expression of one gene in different samples, e.g. comparison of cyt1 in caps and embryos. However, since the efficiency of amplification varies between primer pairs, it is not possible to compare the levels of two different genes. The data shown for each gene is representative of 3-6 independent experiments. The ntl and gsc signals detected in explants at the time of dissection (lane 1) represent less than 5% of the signal in whole embryos. otx2 expression in 6 hour explants (lane 3) averaged about 10% of the expression in whole embryos (see also in situ hybridization in Figs 2C and 5B). (C) Whole-mount in situ hybridization analysis of gene expression in animal cap explants. Each explant shown is an aggregate of 10 animal caps (see Materials and Methods). Explants were analyzed for the expression of gsc (a), ntl (b), pax6 (c), otx2 (d), cyt1 (e) and gta3 (f). A 5 mm section through a cyt1-stained explant (g) shows expression primarily at the surface whereas a section through a gta3 stained explant (h) shows internal expression. The internal staining in g (indicated by an arrow) may surround two cavities. The explants in a were harvested after 2 hours in culture, the explants in d after 6 hours in culture and the explants in b, c, e and f after 20 hours in culture. |
|
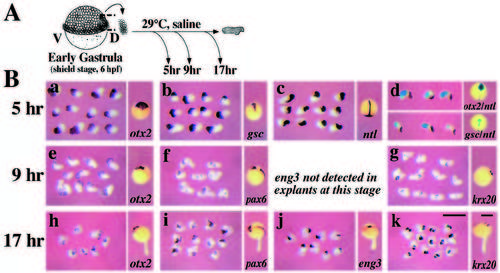
Cultured shields display an anteroposterior arrangement of mesodermal and neural gene expression. (A) Schematic representation of shield dissection and culture. Explants were taken from shield stage embryos (early gastrula, 6 hpf), cultured individually and harvested when control embryos reached the end of gastrulation (10 hpf, 5 hours in culture), the 12 somite stage (15 hpf, 9 hours in culture) or the end of somitogenesis (24 hpf, 17 hours in culture). V,ventral; D, dorsal. (B) Whole-mount in situ hybridization analysis of mesodermal and neural gene expression in explanted shields and whole embryos. After 5 hours in culture, explants (left panel for each marker) and control embryos (right panel) were assayed for expression of otx2 (a), gsc (b), ntl (c) as well as otx2/ntl and gsc/ntl (d; by double in situ hybridization with otx2 (upper panel) or gsc (lower panel) in light blue and ntl in purple). Shields and embryos harvested after 9 hours of culture were assayed for expression of otx2 (e), pax6 (f) and krx20 (g). After 17 hours in culture shields and explants were further assayed for expression of otx2 (h), pax6 (i), eng3 (j) and krx20 (k). The scale bars in (k) are 400 μm. Control embryos are arranged with the anterior up except in d where the embryos are tilted to show the gap in staining between anterior and posterior markers. a-d are dorsal views and e-k are lateral views. (C) Anteroposterior alignment of neural gene expression in cultured shields. Explants hybridized with otx2 (a), pax6 (b) and krx20 (c) after 9 hours in culture were aligned with the posterior knob (arrows, see text) to the right. Each expression domain is indicated with an arrowhead. The relative anteroposterior position of each marker was calculated by determining the distance between the posterior end of the explant and the center of the staining. This distance was expressed as a fraction of the total length of the explant and plotted on the schematic in d. The posterior knob corresponds to an A/P position of ‘0’ and the anterior end to an A/P position of ‘1’. Red dots in d correspond to otx2 expression, blue to anterior pax6, green to krx20, and black to posterior pax6 expression. Each dot represents data from an individual explant. |
|
Anterior neural markers are induced in animal caps following conjugation to shields. (A) Schematic representation of cap/shield conjugate preparation and culture. Each conjugate consisted of five animal caps from sphere stage embryos (late blastula, 4 hpf) fused to a single shield from a shield stage embryo (early gastrula, 6 hpf) and placed in culture (see Material and Methods). Conjugates were harvested when control embryos reached the end of gastrulation (10 hpf, 5 hours in culture), the 12 somite stage (15 hpf, 9 hours in culture) or the end of somitogenesis (24 hpf, 17 hours in culture). D, dorsal; V, ventral. (B) Whole-mount in situ hybridization analysis of mesodermal and neural gene expression in cap/shield conjugates. Conjugates were analyzed for gsc (a) and ntl (b) expression after 5 hours in culture, for pax6 (c), otx2 (d) and krx20 (e) expression after 9 hours in culture and for eng3 (f) expression after 17 hours in culture. The shield lineage label (see Materials and Methods) is visualized in light blue and the animal cap portion is unlabelled. Induction of gene expression in the animal cap portion of the conjugates is indicated by arrowheads. This data is included in Table 2. |
|
Ventral marginal zone explants suppress the expression of otx2 in animal caps. (A) Schematic representation of dissections and conjugate preparation. Animal caps from sphere stage embryos (late blastula, 4 hpf) were aggregated into groups of 10 and either cultured alone or conjugated to ventral marginal zone (VMZ) explants from shield stage embryos (early gastrula, 6 hpf). Each conjugate contained 5 animal caps and 1 VMZ explant. Animal caps and conjugates were harvested when control embryos reached the 12 somite stage (15 hpf, 9 hours of culture). D, dorsal; V,ventral. (B) Whole-mount in situ hybridization analysis of otx2 expression. Animal caps (a) and conjugates (b) were analyzed for otx2 expression after 9 hours in culture. The VMZ lineage label (see Materials and Methods) is visualized in light blue and animal caps are unlabelled. This data is included in Table 3. |