- Title
-
miR-29b-3p Inhibitor Alleviates Hypomethylation-Related Aberrations Through a Feedback Loop Between miR-29b-3p and DNA Methylation in Cardiomyocytes
- Authors
- Wu, F., Yang, Q., Mi, Y., Wang, F., Cai, K., Zhang, Y., Wang, Y., Wang, X., Gui, Y., Li, Q.
- Source
- Full text @ Front Cell Dev Biol

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Correlations between the mRNA expression of |
|
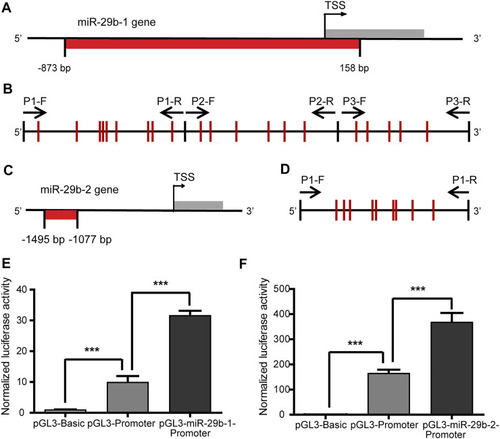
Transcriptional regulatory activity of the miR-29b-1 and miR-29b-2 gene promoters. |
|
Association of miR-29b-3p expression with its methylation status in eight patients with CHD. |
|
The effect of promoter hypermethylation on the expression of miR-29b-3p. |
|
Identification of miR-29b methylation level and mRNA expression after exposure to 5-azacytidine or decitabine. |
|
The effect of miR-29b-3p on the expression of |
|
The impact of miR-29b-3p inhibitor on the overall development of hypomethylated zebrafish embryos. PHENOTYPE:
|
|
The proliferation ability of hypomethylated cardiomyocytes transfected with miR-29b-3p inhibitor. |