- Title
-
Loss of the Bardet-Biedl protein Bbs1 alters photoreceptor outer segment protein and lipid composition
- Authors
- Masek, M., Etard, C., Hofmann, C., Hülsmeier, A.J., Zang, J., Takamiya, M., Gesemann, M., Neuhauss, S.C.F., Hornemann, T., Strähle, U., Bachmann-Gagescu, R.
- Source
- Full text @ Nat. Commun.
|
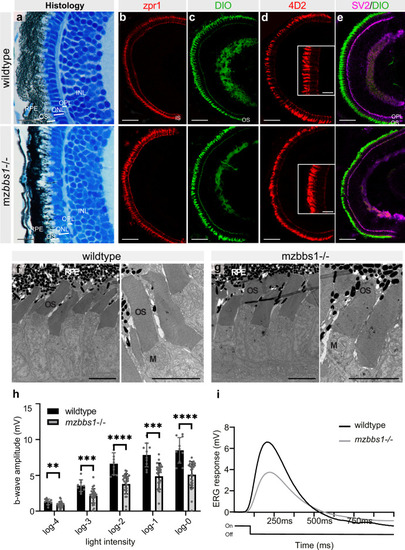
EXPRESSION / LABELING:
PHENOTYPE:
|
|
( |
|
|
|
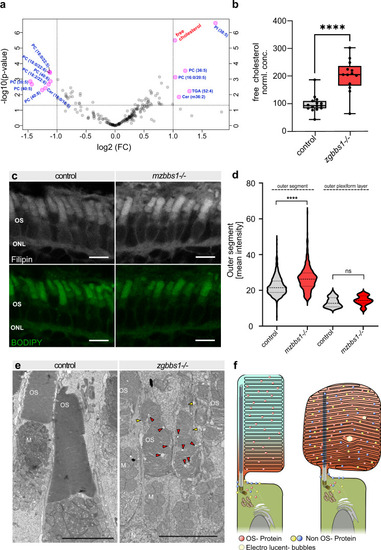
Eye-specific transcriptomic analysis in 5 dpf and 10 dpf maternal zygotic |
|
Quantitative label-free proteomics was applied to mechanically isolated OSs of 5 month old zygotic mutants and sibling controls. |
|
PHENOTYPE:
|