- Title
-
Transcriptomic analysis of zebrafish prion protein mutants supports conserved cross-species function of the cellular prion protein
- Authors
- Pollock, N.M., Leighton, P., Neil, G., Allison, W.T.
- Source
- Full text @ Prion
|
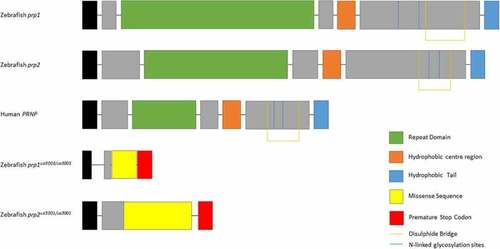
Prion proteins of zebrafish Prp1, Prp2, human PrPC and Prp1 |
|
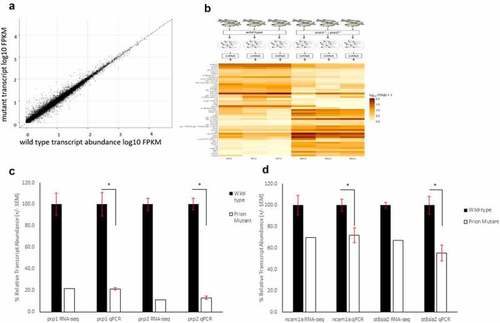
RNA-Sequencing show 1249 genes with an increase or decrease of log2 fold change of 0.5 between wild-type and compound homozygous prion mutant zebrafish larvae. (A) Scatter graph showing relative FPKM values for wild-type (X-axis) and mutant (Y-axis) genes after RNA-sequencing. (B) Methodology diagram showing RNA-sequencing workflow. Two groups, wild-type and prion mutant ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
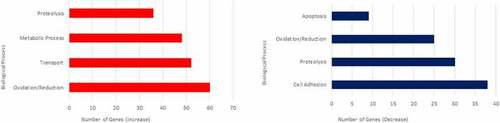
Biological Process Gene Ontologies most affected in 3dpf prion mutant ( |
|
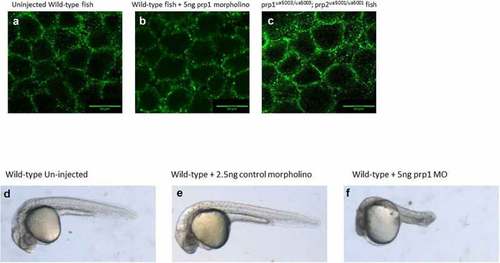
(A–C) There does not appear to be a difference in the maturation or localization of e-cadherin in either 5ng EXPRESSION / LABELING:
PHENOTYPE:
|
|
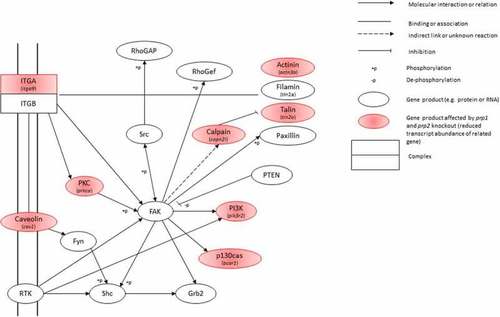
Snapshot of the Focal Adhesion Kinase KEGG pathway. Gene products highlighted in red show those with a significant decrease in transcript abundance, with the specific gene italicized underneath |