- Title
-
Mutation of Gemin5 Causes Defective Hematopoietic Stem/Progenitor Cells Proliferation in Zebrafish Embryonic Hematopoiesis
- Authors
- Liu, X., Zhang, W., Jing, C., Gao, L., Fu, C., Ren, C., Hao, Y., Cao, M., Ma, K., Pan, W., Li, D.
- Source
- Full text @ Front Cell Dev Biol
|
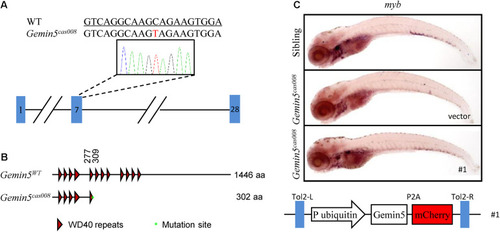
The |
|
The mutation was mapped to a point mutation in gene |
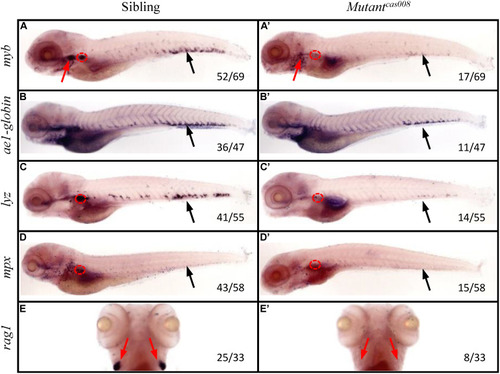
|
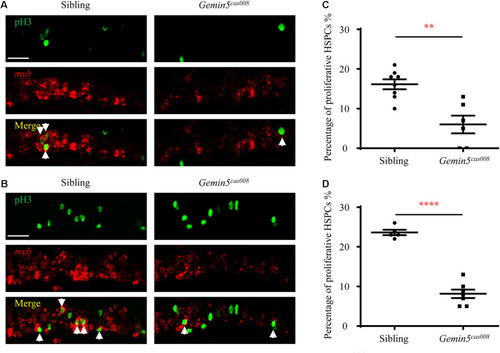
Decreased HSPCs proliferation in mutant |