- Title
-
HIF1α-dependent induction of the mitochondrial chaperone TRAP1 regulates bioenergetic adaptations to hypoxia
- Authors
- Laquatra, C., Sanchez-Martin, C., Dinarello, A., Cannino, G., Minervini, G., Moroni, E., Schiavone, M., Tosatto, S., Argenton, F., Colombo, G., Bernardi, P., Masgras, I., Rasola, A.
- Source
- Full text @ Cell Death Dis.
|
|
|
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
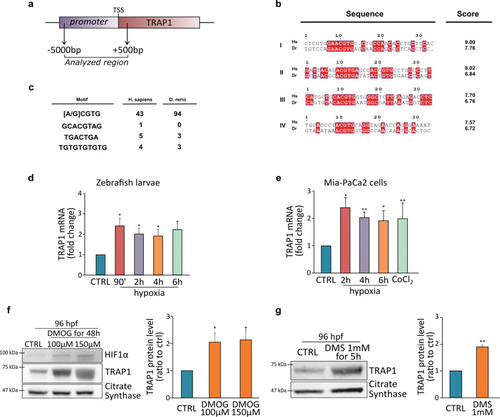
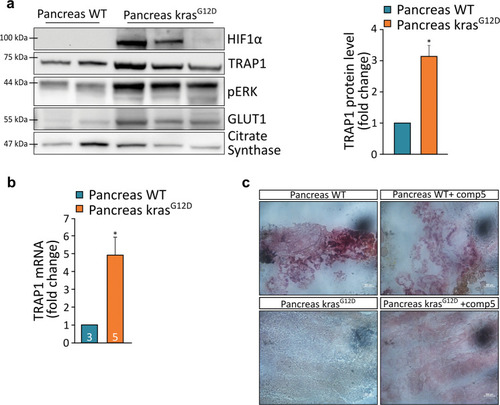
a Schematic representation of the TRAP1 promoter region analyzed that encompasses position −5000 bp to +500 bp from the transcription initiation site. b List of the four (I–IV) HRE regions more conserved between human (Homo sapiens, Hs) and Zebrafish (Danio rerio, Dr) TRAP1 promoters. Scoring was calculated using the position-specific matrix. c Alternative hypoxia-dependent regulative motifs found within the TRAP1 promoter region of Homo sapiens and Danio rerio. d, e Analysis of TRAP1 mRNA expression in wild-type Zebrafish embryos (d) and MIA PaCa-2 human pancreatic adenocarcinoma cells (e) exposed to hypoxia for the indicated times. Values were normalized for expression of rpl13 (d) and actin (e) used as housekeeping genes. f Western blot analysis and protein quantification of TRAP1 expression in embryos at 96 hpf following a 48 h treatment with 100 and 150 µM dimethyloxalyglycine (DMOG). g Western blot analysis and protein quantification of TRAP1 expression in embryos at 96 hpf following a 5 h treatment with 1 mM dimethylsuccinate (DMS). Data are reported as average ±SEM of at least three independent experiments with an unpaired two-tailed Student’s t test; asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001). |
|
|
|
PHENOTYPE:
|