- Title
-
Bmp8a is an essential positive regulator of antiviral immunity in zebrafish
- Authors
- Zhong, S., Li, H., Wang, Y.S., Wang, Y., Ji, G., Li, H.Y., Zhang, S., Liu, Z.
- Source
- Full text @ Commun Biol
|
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
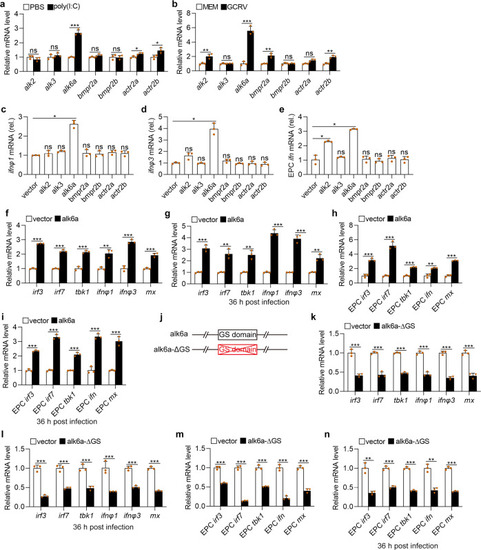
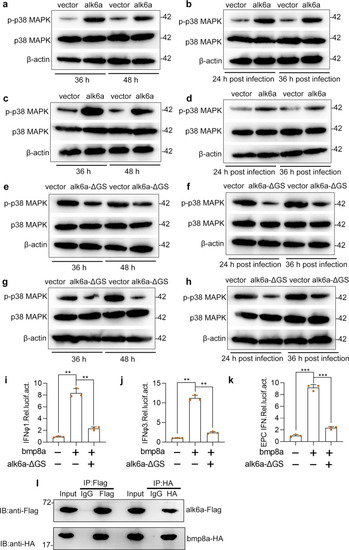
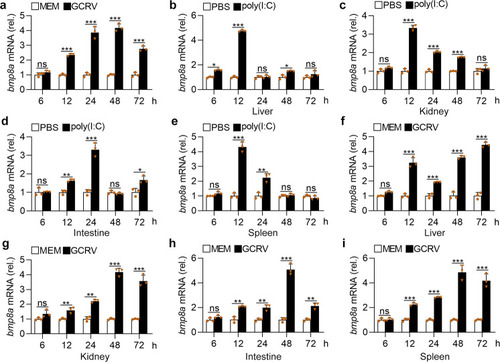
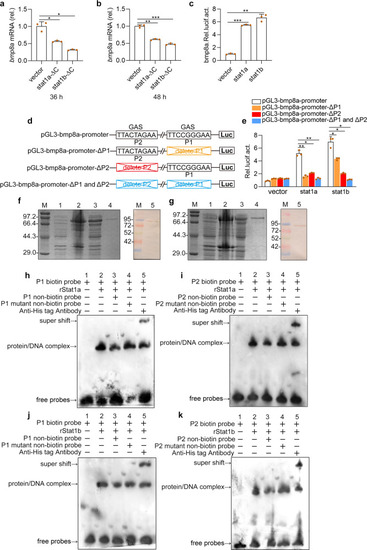
Upon virus infection, the transcriptions of |