- Title
-
Development of a Mucus Gland Bioreactor in Loach Paramisgurnus dabryanus
- Authors
- Zhou, T., Zhou, B., Zhao, Y., Li, Q., Song, G., Zhu, Z., Long, Y., Cui, Z.
- Source
- Full text @ Int. J. Mol. Sci.
|
Construction of a transgenic vector. (A) Plasmid map of the transgenic vector containing a krt8 promoter, grass carp interferon (IFN1) cDNA and His-tag. (B) The expression of EGFP under fluorescence microscope. krt8-1dpf: One day after fertilization; krt8-5dpf: five days after fertilization. (C) Western blot analysis of IFN1 in 293T cells. Group pcDNA-IFN1-His: 293T cells were transfected with pcDNA-IFN1-His; pT2-krt8-IFN1-1, 2 and 3: 293T cells were transfected with pT2-krt8-IFN1; Group pcDNA3.1-His: 293T cells were transfected with pCDNA3.1-His. |
|
Antiviral ability of grass carp IFN1. (A) The pathogenicity of GCRV873 virus particles. The upper bottle contains M199 culture medium supplied with 10% fetal bovine serum; the middle bottle contains M199 culture medium supplied with 10% fetal bovine serum and GCRV873 virus particles at a final concentration of 5.0 × 104 PFU/mL; the lower bottle contains M199 culture medium supplied with 10% fetal bovine serum and GCRV873 virus particles at a final concentration of 5.0 × 105 PFU/mL. (B) The results of crystal violet staining of CIK cells in M199 medium supplied with or without GCRV873 virus particles. 1: CIK cells were infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 2: CIK cells were electrically transfected with pCDNA3.1 and infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 3: CIK cells were electrically transfected with pCDNA-IFN1-His and infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 4: CIK cells were electrically transfected with pCDNA-IFN1-His and not infected with GCRV873. Red arrow: Plaques formed in cultured CIK cells. Yellow arrow: No plaques formed in cultured CIK cells. (C) The transcriptional expression of factors in CIK cells infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL). S5 and S6 are subunits of GCRV873. IFR-9 and STAT1 are key molecules downstream of type I interferon signaling. Data were expressed as mean ± SD. ** p < 0.01; *** p < 0.001. |
|
Generation of transgenic loach. (A) The strategy of generation of transgenic loach. (B) The sensitivity and specificity of seven primer pairs were determined in a 25 μL volume containing 100 ng genomic DNA and transgenic plasmids pT2-krt8-IFN1 (0, 1, 5, 10, 20, 50 or 100 copies) as template. |
|
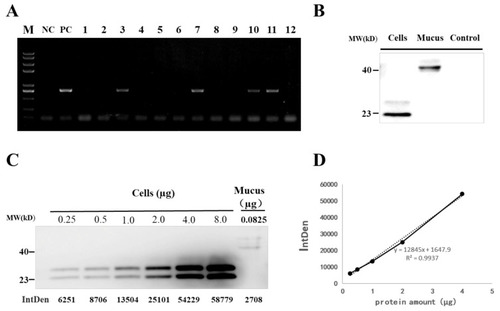
Detection of grass carp IFN1 in transgenic loach. (A) Detection of the IFN1-expressing cassette in F1 offspring by PCR. NC: negative control; PC: positive control; 1–12: F1 loaches. (B) Detection of grass carp IFN1 in mucus using western blots. Cells: the supernatant medium of cultured 293T cells transfected with pcDNA-IFN1-His; Mucus: the mucus collected from F1 loaches containing IFN1-expressing cassette; Control: the mucus collected from wide type loaches. (C) Quantitative analysis of grass carp IFN1 in mucus using western blots. Cells: Purified protein from the supernatant medium of 293T cells transfected with pcDNA-IFN1-His; Mucus: the mucus collected from F1 loaches containing IFN1-expressing cassette. (D) The linear analysis of gray values (IntDen) and protein concentrations in the supernatant medium of 293T cells transfected with pcDNA-IFN1-His to determine the concentration of grass carp IFN1 in the mucus collected from F1 loaches containing IFN1-expressing cassette in (C). |
|
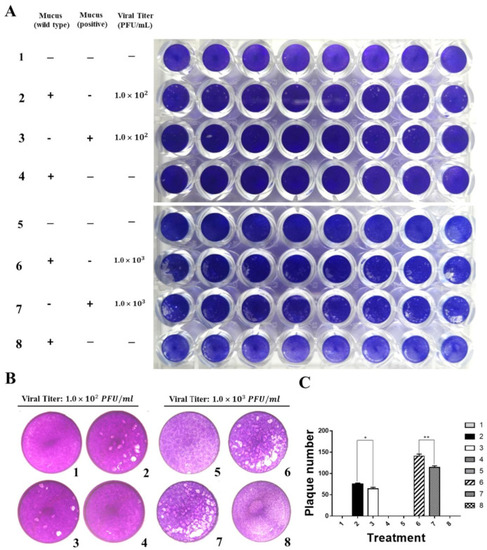
Grass carp IFN1 from mucus inhibited the plaque formation of cultured cells infected with GCRV873. (A,B) Detection of the antiviral ability of grass carp IFN1 from the mucus of transgenic loach. 1, 5: added M199 medium containing 2% FBS; 2: added M199 medium containing 2% FBS and 20% mucus (v/v) from wide type (WT) loaches and infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 3: added M199 medium containing 2% FBS and 20% mucus (v/v) from grass carp IFN1-expressing transgenic loaches and infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 6: added M199 medium containing 2% FBS and 20% mucus (v/v) from WT loaches and infected with GCRV873 (viral titer: 1.0 × 102 PFU/mL); 7: added M199 medium containing 2% FBS and 20% mucus (v/v) from grass carp IFN1-expressing transgenic loaches and infected with GCRV873 (viral titer: 1.0 × 103 PFU/mL); 4, 8: added M199 medium containing 2% FBS and 20% mucus (v/v) from WT loaches. (C) Graph of plaque numbers of the Figure 5A. The experiments were performed in triplicate. Data were expressed as mean ± SD. * p < 0.05; ** p < 0.01. |
|
Analysis of integration site of IFN1-expressing cassette in transgenic loaches. (A) Genomic DNA of grass carp IFN1 cDNA-transgenic loaches was digested with EcoRV and subjected to Southern blotting with digoxigenin-labelled cDNA probes and antidigoxin antibodies. (B) Schematic diagram of the transposon-mediated IFN1-expressing cassette integrated in the genome, showing primer pairs krt8-F1/IFN1-R5 for Southern blots. (C) The electrophoresis of genome walking. AD5 and AD1 are merging primers for the genome of loach. R1, R2, R3 and L1, L2, L3 are primers designed on integrated IFN1-expressing cassette of the plasmid pT2-krt8-IFN1. |