- Title
-
Identification of Potentially Relevant Genes for Excessive Exercise-Induced Pathological Cardiac Hypertrophy in Zebrafish
- Authors
- Zhou, Z., Zheng, L., Tang, C., Chen, Z., Zhu, R., Peng, X., Wu, X., Zhu, P.
- Source
- Full text @ Front. Physiol.
|
Figure 1. Excessive exercise leads to pathological cardiac hypertrophy. (A,B) Representative images from control (A) and excessively-exercised zebrafish hearts (B) after 4 weeks of static and excessive-exercise treatment. (C–F) Representative pictures of Masson’s trichrome-stained ventricular tissues of control (C,D) and excessively-exercised zebrafish hearts (E,F). Blue area indicates fibrosis. (D,F) Are enlargements of the rectangle area in (C,E), respectively. Scale bar = 100 μm. (G) Quantitative analysis of Masson staining positive myocardial fibrosis area using Image J. ∗p < 0.05 by unpaired Student’s t-test. n = 3. (H,I) TEM images of control (H) and excessively exercised heart tissue (I). Scale bar = 5 μm. (J) RT-qPCR analysis of the expression of mitochondrial functional markers. ∗p < 0.05, ∗∗p < 0.01 by unpaired Student’s t-test. (K) Echocardiographic analysis of zebrafish after excessive exercise and in the static control (LVPW, LV posterior wall thickness). Error bars indicate SEM. n = 4. (L) The maximal oxygen consumption (MO2) of zebrafish after excessive exercise and in the control group was analyzed. n = 8. (M) RT-qPCR analysis of the expression of pathologic and physiological hypotrophy-related marker genes. ∗p < 0.05, ∗∗p < 0.01 by unpaired Student’s t-test. |
|
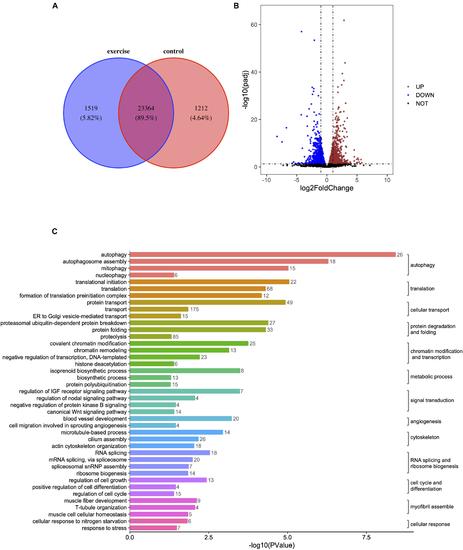
Figure 2. Identification of differentially expressed genes (DEGs) and enrichment analysis of GO terms in zebrafish hearts after excessive exercise and in the control group. (A) Venn diagram of the total number of identified genes in the excessive exercise and control groups. (B) Volcano plot constructed using fold-change values and P adj-values. Red: upregulated, Blue: downregulated. (C) The DEG enrichment analysis of GO terms in the biological process subcategory. The number indicates the number of DEGs that are enriched in the clustered representative terms. |
|
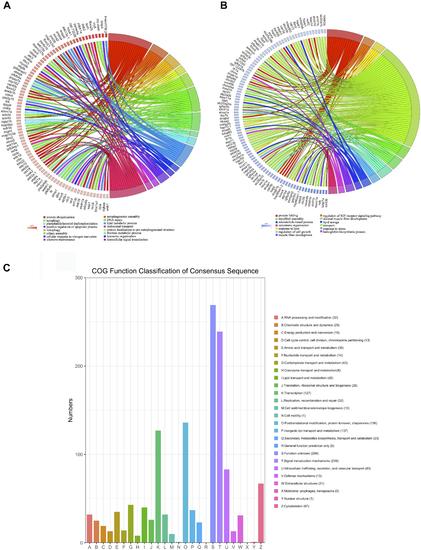
Figure 3. Function analysis of DEGs (p < 0.05, fold change > 2). (A) Upregulated DEGs were assigned to representative biological processes. (B) Downregulated DEGs were assigned to representative biological processes. (C) Cluster of Orthologous Groups (COG) functional classification of the DEGs. The legend shows the name of each function and the proportion of DEGs in each functional class. |
|
Figure 4. RT-qPCR validation of key genes involved in excessive exercise induced pathological cardiac hypertrophy. (A) upregulated genes involved in pathological remodeling. (B) downregulated genes involved in pathological remodeling. n = 8 in control and exercise group. *p < 0.05, **p < 0.01 by unpaired Student’s t-test. |
|
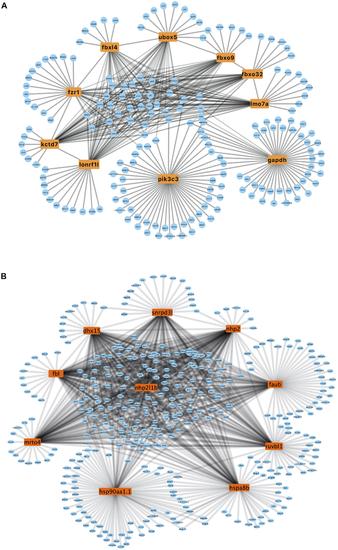
Figure 5. PPI network of upregulated and downregulated genes. (A) PPI network of upregulated hub genes. Hub genes are listed in the box. (B) PPI network of downregulated hub genes. Hub genes are listed in the box. |