- Title
-
Hedgehog signaling regulates neurogenesis in the larval and adult zebrafish hypothalamus
- Authors
- Male, I., Ozacar, A.T., Fagan, R.R., Loring, M.D., Loring, M.D., Shen, M.C., Pace, V.A., Devine, C.A., Lawson, G.E., Lutservitz, A., Karlstrom, R.O.
- Source
- Full text @ eNeuro
|
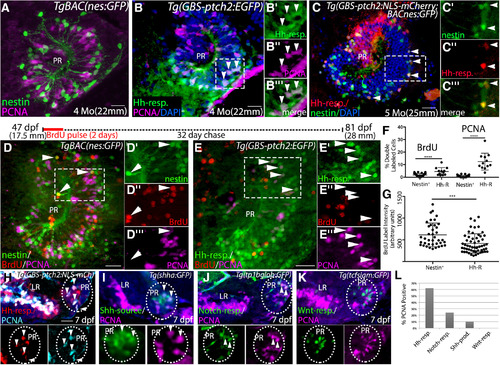
Shh-producing and Hh-responsive radial glial cells in the adult hypothalamus are proliferative neural precursors. |
|
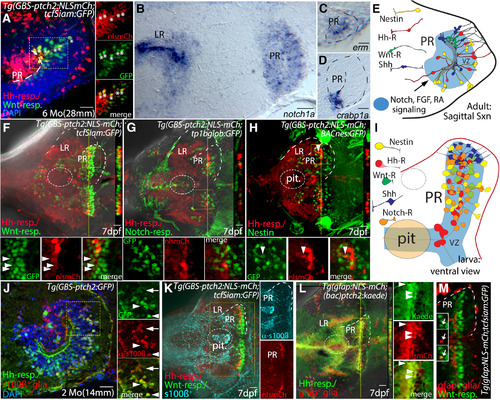
Hh pathway gene expression in the larval and adult zebrafish hypothalamus. |
|
Hh signaling positively regulates proliferation in the adult and larval hypothalamus. |
|
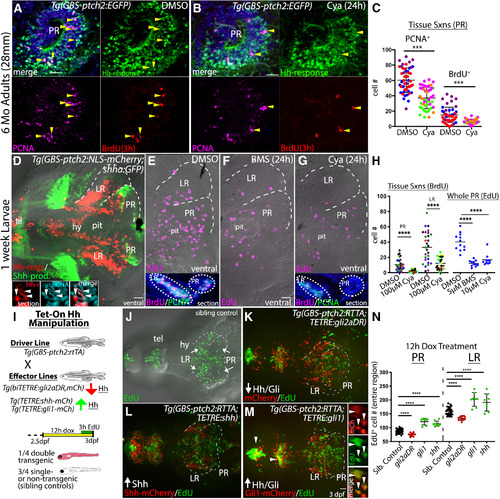
Cya and BMS-833923 both block Hh signaling in zebrafish larvae. |
|
Blocking Hh signaling with Cya reduces |
|
Hh, Wnt, Notch, FGF, and retinoic acid signaling in a complex hypothalamic neurogenic niche. |
|
Hh-responsive cells are more highly proliferative than other radial glia in the hypothalamus. |
|
Hh-responsive progenitors of the hypothalamus give rise to dopaminergic, serotonergic, and GABAergic neurons. |