- Title
-
The endosomal trafficking regulator LITAF controls the cardiac Nav1.5 channel via the ubiquitin ligase NEDD4-2
- Authors
- Turan, N.N., Moshal, K.S., Roder, K., Baggett, B.C., Kabakov, A.Y., Dhakal, S., Teramoto, R., Chiang, D.Y., Zhong, M., Xie, A., Lu, Y., Dudley, S.C., MacRae, C.A., Karma, A., Koren, G.
- Source
- Full text @ J. Biol. Chem.
|
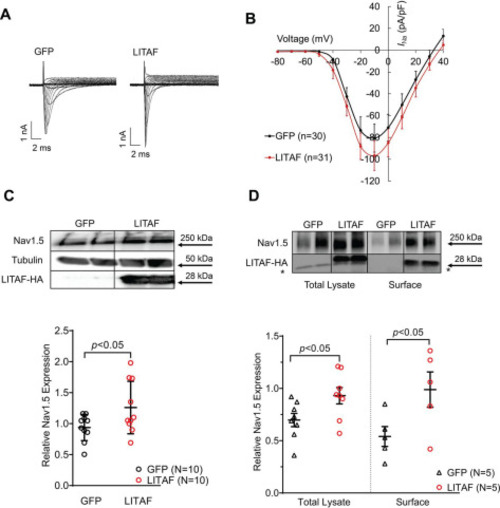
Figure 1. LITAF increases INa and Nav1.5 levels in 3wRbCM. Cardiomyocytes were transduced with adenovirus encoding GFP or LITAF-HA (MOI of 10, 48 h). A, left panel, representative traces of INa in control, GFP-expressing 3wRbCM. INa was activated with depolarizing membrane potentials between −80 and +40 mV from −100 mV holding potential. Right panel, typical INa traces in GFP- and LITAF-expressing 3wRbCM. B, voltage dependence of INa current in transduced cardiomyocytes. The currents were normalized to cell capacitance (means ± S.E.; p < 0.01, 3wRbCM from three rabbits were used for both LITAF and GFP experiments). C, activation curves of INa were obtained from data shown in B, whereas inactivation curves were obtained as described under “Experimental procedures.” D, left panel, protein expression levels of Nav1.5, LITAF-HA, and tubulin. Right panel, respective changes of Nav1.5 protein levels normalized to tubulin. Although the theoretical molecular mass of the HA-tagged human LITAF is 20.6 kDa, our Western blotting data show a specific band at ∼28 kDa, which is likely due to post-translational modification (all values are means ± S.E., Student's t test, n = 11 each). *, p < 0.05. |
|
Figure 2. LITAF increases INa as well as surface expression of Nav1.5 in stable HEK cells. The cells stably co-expressing Nav1.5 and GFP were co-transfected with LITAF- and DsRed-expressing plasmids or control (GFP and DsRed). A, typical Na+ traces in GFP- and LITAF-expressing HEK cells. B, current–voltage relationship for INa. INa was activated from −100 mV holding potential by depolarizing steps up to +40 mV in 10-mV increments. C, top panel, total expression of Nav1.5 and LITAF-HA. Bottom panel, respective change in Nav1.5 expression, normalized to tubulin (all values are means ± S.E.; Student's t test; n = 10). The uncropped probed membrane is shown in Fig. S1. D, total and surface protein expression of Nav1.5 and LITAF. Stable HEK cells were transfected with LITAF or GFP (control) expression plasmids. Cell-surface protein was biotinylated using sulfo-NHS-S-biotin, purified with NeutrAvidin beads from total cell lysates, subjected to SDS-PAGE, and blotted onto a polyvinylidene difluoride membrane. A representative immunoblot shows cell-surface and total lysate expression of Nav1.5, LITAF-HA, and tubulin (top panel) (the asterisk indicates an unspecific band). Respective changes in Nav1.5 expression, normalized to tubulin. All values are means ± S.E. (n = 5) (bottom). |
|
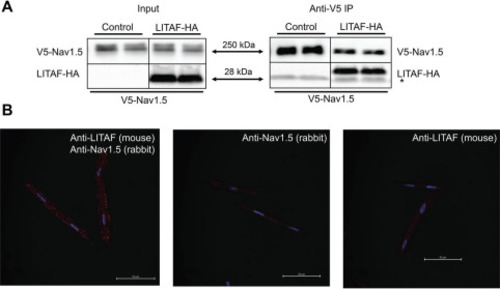
Figure 3. Physical interaction between LITAF and Nav1.5 in HEK cells and 3wRbCM. A, V5 IP of lysates from HEK cells stably expressing V5-tagged Nav1.5 and transfected with plasmids for HA-tagged LITAF or empty expression plasmid (control) (n = 3). The right panel shows immunoprecipitated Nav1.5 and co-precipitated LITAF-HA and thus an interaction between LITAF and Nav1.5 (the asterisk indicates the light chain of the IP capture antibody). Input levels of Nav1.5 and LITAF-HA are shown in the left panel. B, Duolink in situ proximity ligation assay using mouse anti-LITAF and rabbit anti-Nav1.5 antibodies in 3wRbCM. The co-localization between molecules is indicated by red puncta (left panel). Virtually no puncta were detected in negative controls in which only one antibody was used, i.e. rabbit anti-Nav1.5 (middle panel) or mouse anti-LITAF antibodies (right panel). The nuclei were stained with DAPI (blue). Depicted merged confocal images (bright field, DAPI, and Texas Red) are representative of each condition. Scale bar, 50 μm. |
|
Figure 4. LITAF partially abolishes NEDD4-2–dependent down-regulation of INa in HEK cells. A, co-immunoprecipitation of cell lysates from HEK cells transfected with plasmids for NEDD4-2–FLAG and LITAF-HA or just NEDD4-2–FLAG for 48h (n = 3). Top panel, the top part shows an interaction between immunoprecipitated NEDD4-2–FLAG and co-precipitated LITAF-HA (the asterisk indicates the light chain of the IP capture antibody), whereas the bottom part displays input levels of NEDD4-2–FLAG and LITAF-HA. Bottom panel, respective changes in input NEDD4-2 expression, normalized to tubulin. All values are means ± S.E. (n = 3). B, current–voltage relationships of INa peak currents for baseline conditions from cells expressing. GFP (control), NEDD4-2, or NEDD4-2 and LITAF (means ± S.E.). Two-way analysis of variance for repeated measures revealed significant differences in INa between all groups: GFP versus NEDD4-2: F = 80.05, p < 0.0001. NEDD4-2 versus NEDD4-2 + LITAF: F = 11.42, p = 0.0008. GFP versus NEDD4-2 + LITAF: F = 15.28, p = 0.0001. |
|
Figure 5. LITAF down-regulates total NEDD4-2 expression. Adenovirus for expression of GFP (control) or LITAF-HA was incubated with 3wRbCM (10 MOI) (A) or NRbCM (2 MOI) (B) for 48 h. HEK cells were transfected with plasmid encoding GFP (control) or HA-tagged LITAF (C) for 48 h. Representative Western blotting data from respective extracts to measure expression of NEDD4-2, tubulin, and LITAF-HA. D, NEDD4-2–FLAG, LITAF-HA, and tubulin expression in HEK cells transfected with expression plasmids for NEDD4-2–FLAG, LITAF-HA, or control plasmid for 48 h. E, Respective relative LITAF-dependent NEDD4-2 expression levels normalized to tubulin expression from four independent experiments performed in duplicate (Student's t test; p < 0.05). In the scatter plot, averaged values for the four independent experiments together with the means and S.E. values are shown. |
|
Figure 6. LITAF causes ubiquitination and subsequent proteasomal degradation of NEDD4-2 in HEK cells. A, IP of lysates from HEK cells transfected with plasmids for FLAG-tagged NEDD4-2, HA-tagged ubiquitin, FLAG-tagged LITAF, or control plasmid for 48 h was performed with anti-HA antiserum. A representative immunoblot shows levels of ubiquitinated NEDD4-2–FLAG and tubulin and input levels of NEDD4-2–FLAG, LITAF-FLAG, or tubulin. Relative NEDD4-2–FLAG ubiquitination levels as calculated by the ratio of ubiquitinated NEDD4-2 normalized to ubiquitinated tubulin and total NEDD4-2 normalized to total ubiquitin levels (n = 5; means ± S.E.). In the scatter plot, averaged values for the five independent experiments together with the means and S.E. values are shown. Bottom panel, Student's t test, p < 0.01. B, LITAF-mediated degradation of NEDD4-2 through proteasomes. HEK cells were transfected with plasmids for NEDD4-2–FLAG, LITAF-HA or control plasmid for 24 h and then treated with vehicle (▿), 5 μm MG132, or 10 μm chloroquine for 24 h. Top panel, representative Western blots show total expression of NEDD4-2–FLAG, LITAF-HA, and tubulin of treated cells. Bottom panel, respective relative expression levels (means ± S.E.) of total NEDD4-2 normalized to tubulin levels (n = 4, n = 2, p < 0.05). |
|
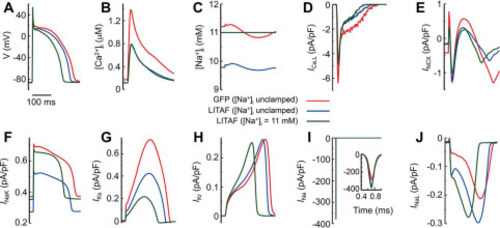
Figure 7. Computer simulations of rabbit ventricular myocytes. Cardiomyocytes were paced at 2.5 Hz under three different conditions: transduced with adenovirus encoding GFP with intracellular sodium concentration [Na+]i unclamped (red traces); adenovirally transduced causing LITAF overexpression with [Na+]i unclamped (blue traces); and adenoviral overexpression of LITAF with [Na+]i clamped to the same average steady-state 11 mm value reached under GFP (green traces). Transmembrane voltage V (A), cytosolic calcium concentration [Ca2+]iB), [Na+]i C), L-type calcium current ICa,L (D), Na+/Ca2+ exchanger current INCX (E), Na+/K+-pump current INaK (F), slowly activating delayed rectifier K+ current IKs (G), rapidly activating delayed rectifier K+ current IKr (H), fast sodium current INa (I), and late sodium current INaL (J). |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |