- Title
-
A novel chemical-combination screen in zebrafish identifies epigenetic small molecule candidates for the treatment of Duchenne muscular dystrophy
- Authors
- Farr, G.H., Morris, M., Gomez, A., Pham, T., Kilroy, E., Parker, E.U., Said, S., Henry, C., Maves, L.
- Source
- Full text @ Skelet Muscle

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
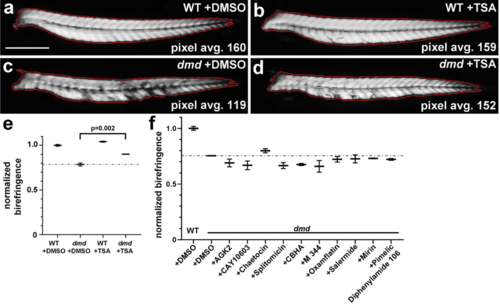
Individual compounds from plate 2 row A do not significantly improve zebrafish dmd mutant muscle birefringence. a-d Four dpf zebrafish trunk muscle, with trunk skeletal muscle birefringence outlined in red. Lateral views, anterior to the left. Average pixel brightness values are shown for (a) wild-type (WT) + DMSO, (b) WT + TSA, (c) dmd + DMSO, and (d) dmd + TSA. Scale bar = 500 μm. e Graph of normalized birefringence pixel intensities for 200 nM TSA treatments vs DMSO controls. n = 3 biological replicates for each treatment. Plot shows the average normalized pixel intensity for the 3 replicates for each treatment (4-9 genotyped animals per replicate). The dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 17). Error bars represent standard error. p value determined by Student’s t test. (f) Graph of normalized birefringence pixel intensities for treatments of zebrafish dmd mutants with the 10 individual chemicals from plate 2 row A. Control treatment is 1% DMSO. All chemicals were tested at 1 μM. For each treatment condition, n = 3 replicates, with 2-9 dmd embryos in each replicate. Plot shows the normalized birefringence pixel intensity for the 3 replicates for each treatment. The dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 15). Error bars represent standard error. Significance was determined using a one-way ANOVA test comparing each treatment group to the dmd DMSO control group with Dunnett’s correction for multiple comparisons. p > 0.2 for all individual chemicals compared to dmd DMSO control |
|
Oxamflatin and salermide mediate the effects of plate 2 row A. a Graph of average pixel intensities for treatments of zebrafish dmd mutants with pilot screen pool #72 and with pool #72 with each individual chemical removed. Control treatment is 1% DMSO. All chemicals included in the pools were tested at 1 μM. For each condition, 3 replicates of 25 embryos each were treated, with 2-9 dmd−/− embryos in each replicate. Plotted are the average normalized pixel intensities of the embryos from all 3 replicates for that treatment (n = 14-26 total dmd−/− embryos per treatment). Treatment with pool #72 without chaetocin, oxamflatin, salermide, or delphinidin chloride did not achieve the rescue effect seen with the full pool #72. The dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 26). Error bars represent standard error. Significance was determined using a one-way ANOVA test comparing each treatment group to the dmd DMSO control group with Dunnett’s correction for multiple comparisons. *p ≤ 0.04 compared to dmd DMSO control. b Graph of average normalized pixel intensities for treatments of dmd mutants with pool #72 and with combinations of chaetocin, oxamflatin, and salermide. Control treatment is 1% DMSO. All chemicals were used at 1 μM. For each treatment condition, n = 3 replicates, with 1-11 dmd−/− embryos in each replicate. Plot shows the average normalized pixel intensity for the 3 replicates for each treatment. The dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 14). Error bars represent standard error. Significance was determined using a one-way ANOVA test comparing each treatment group to the dmd DMSO control group with Dunnett’s correction for multiple comparisons. c-f 4 dpf zebrafish trunk muscle birefringence. Lateral views, anterior to the left. Representative animals are shown from treatments in (b). c WT + DMSO, d dmd + DMSO, e dmd + pool #72, and f dmd + oxamflatin and salermide. Scale bar = 500 μm. g Validation of oxamflatin and salermide treatment effects. Graph of average normalized pixel intensities for treatments of dmd fish with oxamflatin and salermide, performed in the Henry Lab. Treatments were performed from 1-4 dpf. Control treatment is 1% DMSO. Compounds were each used at 1 μM. Dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd−/− animals (n = 11). WT + DMSO, n = 10; WT + ox+sal, n = 9; dmd−/− + ox+sal, n = 14. Error bars represent standard error. p value determined by Student’s t test. h Validation of TSA treatment effects. Graph of average normalized pixel intensities for treatments of dmd fish with TSA, performed in the Henry Lab. Treatments were performed from 1-4 dpf. Control treatment is 1% DMSO. TSA was used at 200 nM. Dashed line represents the average normalized pixel intensity for all of the DMSO-treated dmd−/− animals (n = 31). WT + DMSO, n = 6; WT + TSA, n = 6; dmd−/− + TSA, n = 28. Error bars represent standard error. p value determined by Student’s t test PHENOTYPE:
|
|
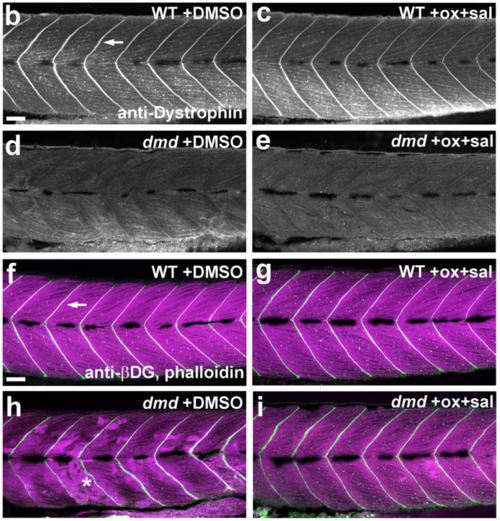
Oxamflatin and salermide have dose-dependent effects, independent of dystrophin expression. a Graph of average normalized pixel intensities for treatments of dmd mutants with doses of oxamflatin and salermide. Control treatment is 1% DMSO. Chemicals were used between 0.5 μM and 4 μM, over two separate experiments. For each treatment condition, n = 4 replicates, with 2-11 dmd−/− embryos in each replicate. Plot shows the average normalized pixel intensity for each of the 4 replicate pools for each treatment. The vertical line separates the treatment conditions from the two experiments, each of which has its own WT + DMSO and dmd + DMSO controls. The dashed lines represent the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 26 and n = 30). Error bars represent standard error. Significance was determined using a one-way ANOVA test comparing each treatment group to the dmd DMSO control group with Dunnett’s correction for multiple comparisons. *p ≤ 0.029, **p = 0.0029, ***p = 0.0007 compared to dmd DMSO control. b-e Confocal images of anti-dystrophin staining in the trunk musculature of 4 dpf b WT + DMSO, c WT + oxamflatin and salermide, d dmd + DMSO, and e dmd + oxamflatin and salermide larvae. Lateral views, anterior to the left. Arrow points to dystrophin expression in the vertical myoseptum. All dmd+/+ animals showed normal dystrophin expression (WT + DMSO, n = 16; WT + ox+sal, n = 14) and all dmd−/− animals lacked detectable dystrophin expression (dmd−/− + DMSO, n = 23; dmd−/− + ox+sal, n = 18). Scale bar = 50 μm. f-i Confocal images of anti-β-dystroglycan (βDG) and phalloidin staining in the trunk musculature of 4 dpf f WT + DMSO, g WT + oxamflatin and salermide, h dmd + DMSO, i dmd + oxamflatin and salermide. Lateral views, anterior to the left. Arrow points to βDG expression (white) in the vertical myoseptum. Phalloidin staining of filamentous actin (magenta) shows the disrupted muscle structure in dmd mutants (* in h). All wild type animals (+/+ and +/−) showed normal β-dystroglycan expression (WT + DMSO, n = 27; WT + ox+sal, n = 26), and dmd−/− animals showed largely maintained β-dystroglycan expression (dmd−/− + DMSO, n = 9; dmd−/− + ox+sal, n = 14). Scale bar = 50 μm |
|
Oxamflatin and salermide have dose-dependent effects, independent of dystrophin expression. a Graph of average normalized pixel intensities for treatments of dmd mutants with doses of oxamflatin and salermide. Control treatment is 1% DMSO. Chemicals were used between 0.5 μM and 4 μM, over two separate experiments. For each treatment condition, n = 4 replicates, with 2-11 dmd−/− embryos in each replicate. Plot shows the average normalized pixel intensity for each of the 4 replicate pools for each treatment. The vertical line separates the treatment conditions from the two experiments, each of which has its own WT + DMSO and dmd + DMSO controls. The dashed lines represent the average normalized pixel intensity for all of the DMSO-treated dmd animals (n = 26 and n = 30). Error bars represent standard error. Significance was determined using a one-way ANOVA test comparing each treatment group to the dmd DMSO control group with Dunnett’s correction for multiple comparisons. *p ≤ 0.029, **p = 0.0029, ***p = 0.0007 compared to dmd DMSO control. b-e Confocal images of anti-dystrophin staining in the trunk musculature of 4 dpf b WT + DMSO, c WT + oxamflatin and salermide, d dmd + DMSO, and e dmd + oxamflatin and salermide larvae. Lateral views, anterior to the left. Arrow points to dystrophin expression in the vertical myoseptum. All dmd+/+ animals showed normal dystrophin expression (WT + DMSO, n = 16; WT + ox+sal, n = 14) and all dmd−/− animals lacked detectable dystrophin expression (dmd−/− + DMSO, n = 23; dmd−/− + ox+sal, n = 18). Scale bar = 50 μm. f-i Confocal images of anti-β-dystroglycan (βDG) and phalloidin staining in the trunk musculature of 4 dpf f WT + DMSO, g WT + oxamflatin and salermide, h dmd + DMSO, i dmd + oxamflatin and salermide. Lateral views, anterior to the left. Arrow points to βDG expression (white) in the vertical myoseptum. Phalloidin staining of filamentous actin (magenta) shows the disrupted muscle structure in dmd mutants (* in h). All wild type animals (+/+ and +/−) showed normal β-dystroglycan expression (WT + DMSO, n = 27; WT + ox+sal, n = 26), and dmd−/− animals showed largely maintained β-dystroglycan expression (dmd−/− + DMSO, n = 9; dmd−/− + ox+sal, n = 14). Scale bar = 50 μm |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|