- Title
-
VBP1 modulates Wnt/β-catenin signaling by mediating the stability of the transcription factors TCF/LEFs
- Authors
- Zhang, H., Rong, X., Wang, C., Liu, Y., Lu, L., Li, Y., Zhao, C., Zhou, J.
- Source
- Full text @ J. Biol. Chem.
|
Figure 1. Overexpression of VBP1 inhibits Wnt/β-catenin signaling. A, TOPFlash luciferase assays in HEK293T cells with 500-ng FLAG-tagged VBP1 plasmid transfection. Expression of FLAG-VBP1 was confirmed by Western blotting. Data are from four independent experiments with individual data points shown. B, relative mRNA levels of indicated Wnt target genes in control or VBP1-overexpressing HEK293T cells were analyzed by qRT-PCR. Data are from three independent experiments with individual data points shown. C, TOPFlash luciferase assays in zebrafish embryos with 600-pg VBP1 mRNA injection. Data are from three independent experiments with individual data points shown. D, TOPFlash luciferase assays in Wnt3a-, BIO-, β-CatΔN-, or VP16-Tcf7l1ΔN-treated HEK293T cells with VBP1 overexpression. Control or VBP1-overexpressing HEK293T cells were cotransfected with Renilla and TOPFlash plasmids. The activation of Wnt/β-catenin signal was induced by BIO (1 µm) treatment or by indicated plasmid DNAs (15 ng of Wnt3a, 30 ng of β-CatΔN, or 50 ng of VP16-Tcf7l1ΔN) transfection. Data are from three independent experiments with individual data points shown. E, TOPFlash luciferase assays in wnt3a- or vp16-tcf7l1ΔN-injected zebrafish embryos with VBP1 overexpression. TOPFlash and Renilla plasmids were co-injected into embryos with gfp or VBP1 mRNA. The activation of Wnt/β-catenin signal was induced by injecting 100 pg of wnt3a mRNA or 50 pg of vp16-tcf7l1ΔN mRNA. Data are from three independent experiments with individual data points shown. F, overexpression of VBP1/vbp1 antagonized wnt3a-, β-catΔN-, or vp16-tcf7l1ΔN-induced dorsalized phenotype in zebrafish embryos. Lateral view of representative injected embryos at 5-somite stage was shown. Scale bar, 200 µm. G–I, quantitation of embryos displaying normal or dorsalized phenotype in F. Data are from three independent experiments, and the embryo number of each group is shown on the top. The values are mean ± S.D. Unpaired t test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, nonsignificant. |
|
Figure 2. VBP1 is required for Wnt/β-catenin signaling activity. A, VBP1 protein levels in stable shRNA-treated HEK293T and U2OS cells. Densitometric analysis was from three independent experiments with individual data points shown. B and C, the transcriptional levels of indicated Wnt target genes in control or VBP1-depleted HEK293T (B) and U2OS (C) cells were analyzed by qRT-PCR. Data are from three independent experiments with individual data points shown. D, representative images of injected embryos. GFP fluorescence was detected at the midbrain in TOPdGFP transgenic embryos at 24 hpf. The pattern of otx2 expression did not change in the corresponding injected embryos. Scale bar, 200 µm. E, quantification of green fluorescence in (D) as relative fluorescence intensity. Image J software was used to measure the fluorescence intensity. Data are from three independent experiments, and the embryo number of each group is shown on the top. F, Wnt/β-catenin activity in Wnt3a-, LiCl-, β-CatΔN-, and VP16-Tcf7l1ΔN-treated cells with VBP1 knockdown. TOPFlash plasmid was cotransfected with Renilla plasmid into control or VBP1-knockdown cells. Wnt activity was induced by LiCl (30 mm) or indicated plasmid DNAs (10 ng of Wnt3a, 15 ng of β-CatΔN, or 30 ng of VP16-Tcf7l1ΔN). Data are from three independent experiments with individual data points shown. G, TOPFlash luciferase assays in shRNA-expressed U2OS (left) or HEK293T cells (right). Wnt/β-catenin activity was rescued by a shRNA-resistant zebrafish Vbp1. The activation of Wnt/β-catenin signal was induced by LiCl treatment (left) or transfection with β-CatΔN (right) plasmid DNA. Data are from three independent experiments with individual data points shown. The values are mean ± S.D. Unpaired t test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. One-way analysis of variance followed by Tukey's post-test. Groups labeled with different letters are significantly different from each other. |
|
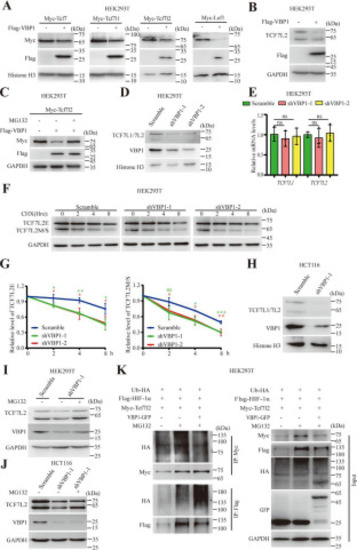
Figure 3. Overexpression or knockdown of VBP1 promotes protein degradation of TCF/LEFs. A, exogenous Tcf/Lef protein levels in control or VBP1-overexpressing HEK293T cells. Similar results were obtained from four experiments. B, endogenous TCF7L2 protein level in control or VBP1-overexpressing HEK293T cells. Similar results were obtained from three experiments. C, changes in exogenous TCF7L2 protein level in VBP1-overexpressing HEK293T cells treated with MG132. Cells transfected with indicated plasmid DNA and treated with or without 10 µm MG132 for 8 h. Similar results were obtained from three experiments. D, TCF7L1 and TCF7L2 protein levels in control or VBP1-knockdown HEK293T cells. Similar results were obtained from three experiments. E, qRT-PCR analysis of transcriptional levels of TCF7L1 and TCF7L2 in control and VBP1-knockdown HEK293T cells. Data are from three independent experiments with individual data points shown. F, representative image of TCF7L2 protein levels in control or VBP1-knockdown HEK293T cells treated with CHX in a time series. G, quantitation of relative protein levels from the two isoforms of TCF7L2, TCF7L2E (left panel) and TCF7L2M/S (right panel), in (F), respectively. The band intensity was quantified using ImageJ software. Data are from three independent experiments. H, TCF7L1 and TCF7L2 protein levels in control or VBP1-knockdown HCT116 cells. Similar results were obtained from three experiments. I, changes in TCF7L2 protein levels in VBP1-knockdown HEK293T cells treated with MG132. VBP1-depleted HEK293T cells treated with or without 10 µm MG132 for 8 h. Similar results were obtained from three experiments. J, changes in TCF7L2 protein levels in VBP1-knockdown HCT116 cells treated with MG132. VBP1-knockdown HCT116 cells treated with or without 10 µm MG132 for 8 h. Similar results were obtained from three experiments. K, ubiquitylation assays in HEK293T cells transfected with indicated plasmids. HIF-1α was used as a positive control of ubiquitylation promoted by VBP1. Similar results were obtained from three experiments. The values are mean ± S.D. Unpaired t test. *p < 0.05; **p < 0.01; ***p < 0.001; ns, nonsignificant. |
|
Figure 4. VBP1 interacts with TCF/LEFs and pVHL. A, the interaction between exogenous TCF/LEFs and VBP1. HEK293T cells were cotransfected with FLAG-tagged VBP1 and Myc-Vector, Myc-Tcf7, Myc-Tcf7l1, Myc-Tcf7l2, or Myc-Lef1, respectively. After 48 h, the cell extracts were immunoprecipitated with an anti-Myc antibody. The immunoprecipitates and the inputs were analyzed by Western blotting with indicated antibody. Similar results were obtained from three experiments. B, the interaction among exogenous VBP1, endogenous TCF7L2, and pVHL. HEK293T cells were transfected with FLAG-tagged VBP1. After 48 h, the cell extracts were immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates and the inputs were analyzed by Western blotting with indicated antibody. Similar results were obtained from three experiments. C, VBP1 bound directly to TCF/LEF. GST and GST-VBP1 proteins expressed by bacteria were incubated with GSH beads and the extracts from nontransfected HEK293T cells (for endogenous TCF7L2 detection) or HEK293T cells transfected with Myc-Tcf/Lefs or FLAG-pVHL. Red asterisk indicates the specific band of GST-VBP1. Similar results were obtained from three experiments. D, BiFC assays detected the binding of VBP1 to Tcf7l1/Tcf7l1ΔN/Tcf7l1ΔNLS in HeLa cells. HeLa cells were cotransfected with FLAG-VBP1-VC and Myc-VN-Tcf7l1, Myc-VN-Tcf7l1ΔN, or Myc-VN-Tcf7l1ΔNLS, respectively. After 24h, Venus in green indicates the interaction between VN-tagged and VC-tagged proteins. Tcf7l1/Tcf7l1ΔN/Tcf7l1ΔNLS was detected by an anti-Myc antibody (red). Nucleus was stained with 4′,6-diamidino-2-phenylindole (blue). Scale bar, 10 µm. Similar results were obtained from three experiments. E and F, mapping of the region in TCF7L2 responsible for the VBP1 interaction. Schematic diagram of human TCF7L2 protein domains is shown in (E). Various HA-tagged TCF7L2 deletion mutants were cotransfected with FLAG-tagged VBP1 in HEK293T cells and cell lysates were subjected to co-immunoprecipitation (F). Red asterisk indicates the specific band. Similar results were obtained from three experiments. |
|
Figure 5. VBP1 regulates TCF/LEFs protein stability through pVHL. A, TCF7L2 protein levels in control or VBP1-overexpressing 786-O cells. Similar results were obtained from three experiments. B, TCF7L2 protein levels in control or VBP1-knockdown 786-O cells. Similar results were obtained from three experiments. C, the association between endogenous TCF7L2 and pVHL in control or VBP1-overexpressing cells. HEK293T cells were transfected with FLAG-tagged VBP1. After 48 h, the cell extracts were immunoprecipitated with an anti-TCF7L2 antibody. The immunoprecipitates and the inputs were analyzed by Western blotting with indicated antibody. D and E, the association between endogenous TCF7L2 and pVHL in control or VBP1-knockdown HEK293T (D) and HCT116 (E) cells. Cell extracts were immunoprecipitated with an anti-TCF7L2 antibody. The immunoprecipitates and the inputs were analyzed by Western blotting with indicated antibody. Similar results were obtained from three experiments. F and G, the transcriptional levels of indicated Wnt target genes in VBP1-overexpressing (F) or -depleted (G) HCT116 cells were analyzed by qRT-PCR. FLAG-VBP1 or endogenous VBP1 protein levels in VBP1-overexpressing or -depleted HCT116 cells were analyzed by Western blotting. Data are from 3–5 independent experiments with individual data points shown. H, representative images of colony formation assay were shown. I, quantification of the colony number shown in (H). Data are from four independent experiments with individual data points shown. Values are means ± S.D. Unpaired t test, two-tailed. *p < 0.05; **p < 0.01; ns, nonsignificant. |