- Title
-
Camel regulates development of the brain ventricular system
- Authors
- Yang, S., Emelyanov, A., You, M.S., Sin, M., Korzh, V.
- Source
- Full text @ Cell Tissue Res.
|
The comparison of Camel to other proteins of L1-CAM family of proteins and |
|
The expression pattern of EXPRESSION / LABELING:
|
|
The expression pattern of EXPRESSION / LABELING:
PHENOTYPE:
|
|
The expression pattern of EXPRESSION / LABELING:
|
|
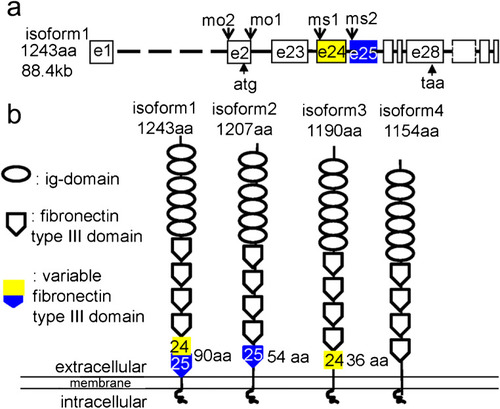
The schematics of organization of the camel genomic DNA and four differentially spliced isoforms. The putative proteins encoded by these mRNA isoforms vary at the level of the fourth fibronectin type III domain encoded by exons 24 and 25. a Organization of camel genomic DNA showing the target sites for morpholino and color-coded exons 24 (yellow) and 25 (blue). b Organization of the putative camel isoforms. The two halves of the sixth fibronectin type III domain are color-coded according to a. The Camel domain structure is generated using this software (http://expasy.org/prosite) |
|
|
|
PHENOTYPE:
|
|
Rescue of hydrocephalus in Camel morphants by |
|
Camel required for the development of SCO and formation of Reissner fiber. |
|
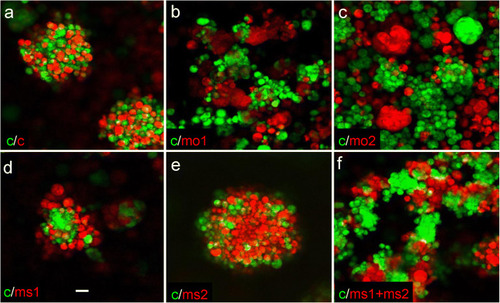
Pan-Camel and isoform-specific anti-Camel morpholino–mediated LOF differentially affects the formation of RF. |
|
The overexpression of |
|
Schematics show organization of the Reissner fiber in respect of the ventricular system (based on Figs. |

Unillustrated author statements |