- Title
-
Generation and characterization of keap1a- and keap1b-knockout zebrafish
- Authors
- Nguyen, V.T., Bian, L., Tamaoki, J., Otsubo, S., Muratani, M., Kawahara, A., Kobayashi, M.
- Source
- Full text @ Redox Biol.
|
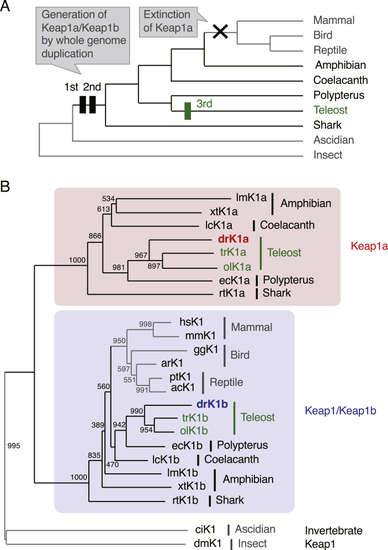
(A) Simplified phylogeny of vertebrate and invertebrate animals. Black and green rectangles represent two WGD events that occurred early in vertebrate evolution and an additional WGD in the teleost ancestor, respectively. (B) Phylogenetic tree of Keap1 family proteins. Amino acid sequences in the broad complex, tramtrack and bric-a-brac domains–intervening region (BTB–IVR) were obtained from GenBank ( |
|
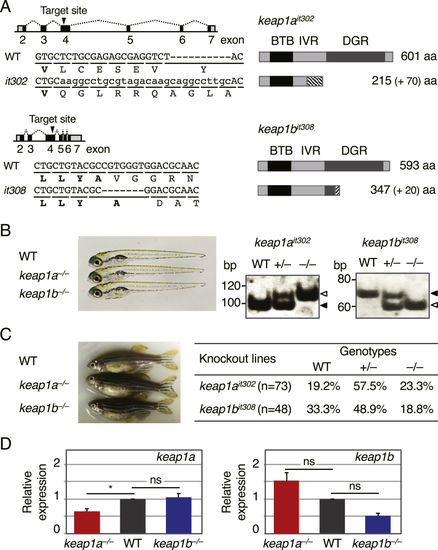
(A) Gene knockout of (B) (C) (D) The relative expression of |
|
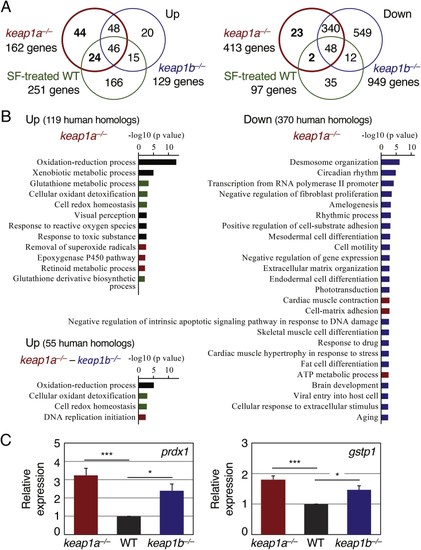
(A) Venn diagrams showing up- (left) and down-regulated genes (right) in 5-dpf larvae of (B) A GO enrichment analysis of biological processes in (C) The comparison of the basal expression of Nrf2 target genes between wild-type and Keap1 knockout larvae. The expression of |
|
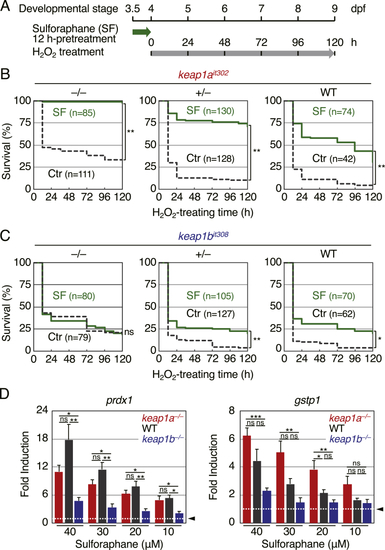
(A) The survival rate of (B) The survival rate of PHENOTYPE:
|
|
(A) A schematic illustration of the experiment. Larvae were treated with 2 mM of H2O2 at 4 dpf after 12-h pretreatment with 40 μM sulforaphane (SF). (B, C) The survival rate of (D) The fold increase in the expression of Nrf2 target genes in |