- Title
-
RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether
- Authors
- Liao, Y.C., Fernandopulle, M.S., Wang, G., Choi, H., Hao, L., Drerup, C.M., Patel, R., Qamar, S., Nixon-Abell, J., Shen, Y., Meadows, W., Vendruscolo, M., Knowles, T.P.J., Nelson, M., Czekalska, M.A., Musteikyte, G., Gachechiladze, M.A., Stephens, C.A., Pasolli, H.A., Forrest, L.R., St George-Hyslop, P., Lippincott-Schwartz, J., Ward, M.E.
- Source
- Full text @ Cell
|
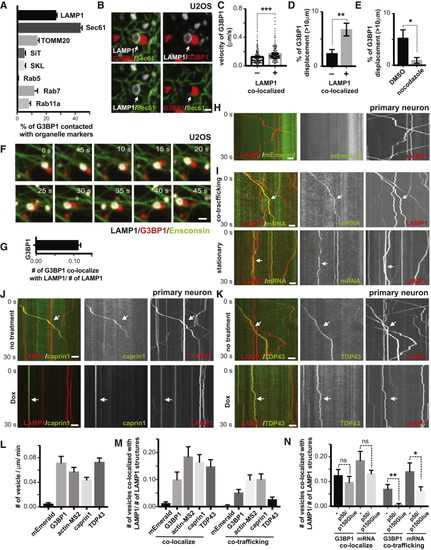
RNA Granules Hitchhike on Motile Lysosomes in Mammalian Cells (A) RNA granule co-imaging with different organelles. U2OS cells expressing mCherry-G3BP1 and different organelle markers were imaged live 30 minutes after heat shock (43oC). Organelle markers: LAMP1 – lysosome, Sec61 – ER, TOMM20 – mitochondria, SiT – Golgi, SKL – peroxisome, Rab5 – early endosome, Rab7 – late endosome, Rab11a – recycling endosome, Ensconsin – microtubule. Arrows point to lysosome-RNA granule contact sites. Scale bar: 2μm. See also (B) Percentage of RNA granules that co-traffic with different organelles from ( (C) Time-lapse image sequence showing RNA granule (mCherry-G3BP1) co-trafficking with a lysosome (LAMP1-HaloTag) along a microtubule (Ensconsin-GFP) in U20S cells immediately after heat shock at 43oC. Scale bar: 1μm. See also (D) Kymograph of RNA granules co-trafficking with lysosomes in axons. Axons of rat cortical neurons expressing LAMP1-HaloTag and mEmerald-G3BP1 were imaged at 100ms/frame for 30 seconds. Arrow points to a lysosome co-trafficking with a G3BP1-labeled structure. P50/p150Glued: Doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin was used to inhibit motor-directed transport of lysosomes. Scale bar: 5 μm. See also (E) Kymograph of mRNA co-trafficking with lysosomes in axons. Axons of rat cortical neurons expressing LAMP1-HaloTag, actin-24xMBS and MCP-NLS-2xEGFP were imaged as in (D). Arrow points to a lysosome co-trafficking with actin mRNA. Scale bar: 5 μm. See also (F) CLEM images of an RNA granule associated with a lysosome. Upper panel shows the fluorescent image of a LAMP1-labeled lysosome and a G3BP1-labeled RNA granule. Lower panel shows the correlated electron microscopy image. L, lysosome; G, RNA granule. Scale bar: 1μm. |
|
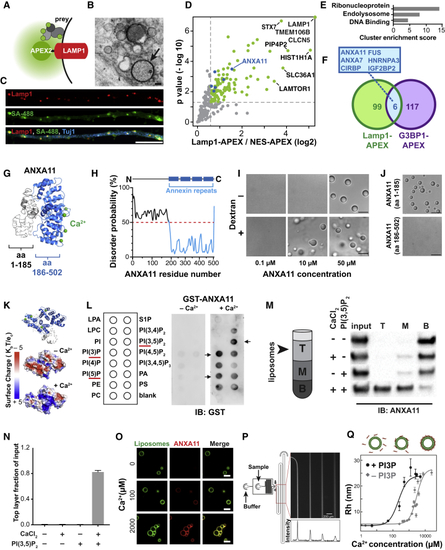
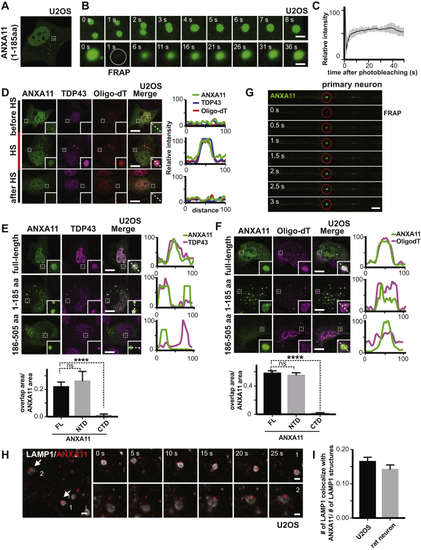
Identification of ANXA11 as a Potential Mediator of RNA Granule-Lysosome Associations (A–G) Proximity labeling proteomic screen for lysosomal interacting proteins in i3Neurons. (A) Schematic of LAMP1-APEX2 bait. (B) Electron microscopy image of DAB precipitate generated by LAMP1-APEX2 (dark contrast, arrow) surrounding lysosomes in i3Neurons. Scale bar: 100 nm. (C) Confocal immunofluorescence image of LAMP1-APEX2 biotinylated prey (streptavidin-488 staining) surrounding LAMP1-positive lysosomes in i3Neuron axons (Tuj1). Scale bar: 10 μm. (D) Plot showing statistically significant LAMP1-APEX2 enriched prey proteins from proximity-labeling proteomics in i3Neurons. n = 4, p values corrected for multiple comparisons. (E) Functional Annotation Clustering of DAVID Gene Ontology terms of Lamp1-APEX enriched prey. (F) Venn diagram of LAMP1-APEX2 hits versus G3BP1-APEX2 stress-granule hits ( (G) Predicted structural analysis of ANXA11 revealed four C-terminal calcium-binding annexin repeats (blue), and a disordered N-terminal region. (H–J) Recombinant ANXA11 undergoes liquid-liquid phase separation (H) PrDOS analysis of ANXA11 predicted a high likelihood of disorder of aa 1-185. (I) Full-length ANXA11 formed spherical, fusing liquid droplets at concentrations above 50μM (upper panel). Phase separation of ANXA11 was facilitated by 10% dextran, with phase separation occurring at lower ANXA11 concentrations (≥ 10μM). Scale bar: 5 μm. See also (J) The disordered N-terminus (aa 1-185, upper panel) of ANXA11 but not C-terminus (aa 186-502, lower panel) underwent liquid-liquid phase separation. Scale bar: 5 μm. (K–Q) Recombinant ANXA11 interacts with negatively charged lysosome-associated phospholipids. (K) Surface maps of predicted ANXA11 structure +/– Ca2+ showing increased positive surface charge (blue) in the presence of Ca2+. Top panel shows orientation of ANXA11 and location of Ca2+ ions (green). (L) Protein lipid overlay assay of recombinant ANXA11-GST protein with membrane lipids. Recombinant ANXA11-GST protein was incubated with a membrane lipid strip +/– Ca2+, followed by anti-GST immunoblotting. Arrowheads indicate enriched lipid binding. Red line highlighted the correlated phospholipid species. (M) Liposome flotation assay of recombinant ANXA11 with liposomes containing PI(3,5)P2 in the absence or presence of Ca2+. Liposomes with associated proteins floated to the top layer following ultra-centrifugation (schematic). ANXA11 in the top (T), middle (M) and bottom (B) fractions was detected via anti-ANXA11 western blot. (N) Quantification ANXA11 enrichment in the top liposome fraction in (O) Microscopy analysis of calcium-dependent recruitment of recombinant ANXA11 protein to fluorescent PI3P-containing liposomes. Representative images showed ANXA11 binding to PI3P-containing liposomes at the indicated calcium concentrations. Scale bar, 5μm. (P) Microfluidic device design for diffusional sizing assay of calcium-dependent ANXA11 binding to liposomes. Inset indicates detection area. The fluorescence intensity along the channel indicates different diffusion times of ANXA11. Scale bar, 200 μm. (Q) Microfluidic diffusional sizing assay to assess changes in molecular radius of ANXA11 upon Ca2+-dependent binding to liposomes (top panel). Bottom panel: Quantification of hydrodynamic radius of ANXA11 when binding to liposomes with (dots) and without (diamonds) PI3P versus Ca2+ concentration. Data were fitted with a Hill binding model. |
|
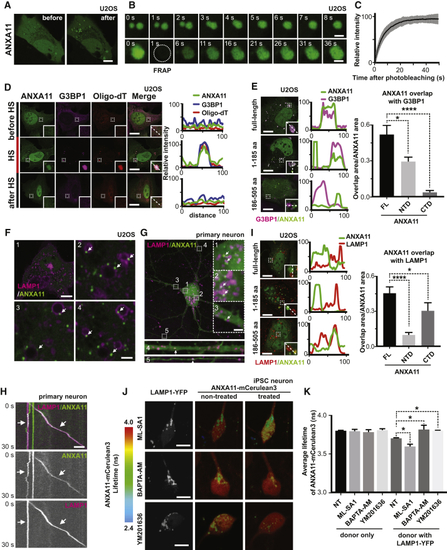
ANXA11 Interacts with Both RNA Granules and Lysosomes in Living Cells (A–D) ANXA11 interact with RNA granules in cells (A) ANXA11-mEmerald redistributes from the cytoplasm into dispersed puncta immediately following heat shock (43oC) in U2OS cells. Scale bar: 20 μm. See also (B) Heat shock induced ANXA11-mEmerald puncta in U2OS cells are motile and undergo fusion (upper panel), and recover rapidly after photobleaching (i.e., FRAP)(bottom panel). Scale bar: 1 μm. See also (C) Quantification of FRAP experiment in (B), n=23. Error bars = SEM. See also (D) Immunostaining of mEmerald-tagged ANXA11 with RNA granule markers (Cy3-Oligo dT(30), anti-G3BP1) before, during and 4 hours after heat shock (HS) in U2OS cells. Line scans show the related intensity profiles of ANXA11 with mRNA (Cy3 Oligo-dT) and with G3BP1. Scale bar: 30 μm. See also (E) Immunostaining of mEmerald-tagged ANXA11 full-length, N-terminal or C-terminal domain and G3BP1 following 30 minutes of heat shock (43oC) in U2OS cells. Line scans show the related intensity profiles of ANXA11 with G3BP1. Scale bar: 30 μm. Right panels show the quantification of ANXA11 tructation area overlap with G3BP1(relative to ANXA11 area). One-way ANOVA, ∗p < 0.05, ∗∗∗∗p < 0.0001, n=10. Error bars = SEM. Scale bar: 30 μm. See also (F–K) ANXA11 puncta interact with lysosomes in cells (F) Panel 1: Live cell imaging of U2OS cells expressing ANXA11-mEmerald and LAMP1-HaloTag following heat shock at 43oC in U2OS cells. Panels 2-4: Enlarged areas of panel 1, with arrows pointing to ANXA11/lysosome contact sites (Scale bar: 1 μm). See also (G) Rat cortical neurons expressing LAMP1-HaloTag and ANXA11-mEmerald imaged after heat shock. ANXA11 puncta co-localized with lysosomes in different neuronal regions: soma (1,2), dendrite (3,4) and axon (5). Arrows point to contact sites between lysosomes and ANXA11 puncta. Scale bar: 5 μm (H) Live imaging of rat cortical neuron axons expressing LAMP1-HaloTag and ANXA11-mEmerald. The corresponding kymograph shows ANXA11 puncta (green) either co-trafficking or co-localizing with lysosomes (magenta). Scale bar: 5 μm. See also (I) Immunostaining of mEmerald-tagged ANXA11 full-length, N-terminal or C-terminal domain with LAMP1-positive lysosomes in U2OS cells following 30 minutes of heat shock (43oC). Line scans show related intensity profiles of ANXA11 and LAMP1. Scale bar: 30 μm. Far right panel shows the quantification of ANXA11 trucations and LAMP1 co-localization (relative to ANXA11 area). One-way ANOVA, ∗p < 0.05, ∗∗∗∗p < 0.0001, n=10. Error bars = SEM. Scale bar: 30 μm. (J) FLIM-FRET analysis of the interaction between ANXA11 and lysosomes and its regulation by lysosomal Ca2+ and PI(3,5)P2. Human i3Neurons were transduced with ANXA11-mCerulean3 (FRET donor) and LAMP-YFP (FRET acceptor). FLIM-FRET images were acquired for the same neurons before and after treatment with ML-SA1, BAPTA-AM or YM201636, and the lifetime of the ANXA11-mCerulean3 signal was determined. Left vertical panels show intensity images of LAMP1-YFP with the various treatments. Middle and right panels show ANAX11-mCeurlean3 lifetimes before and after drug treatment. (K) Quantification of FLIM-FRET lifetime measurements from ( |
|
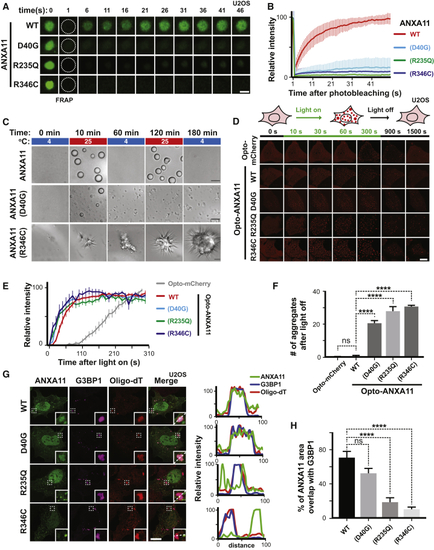
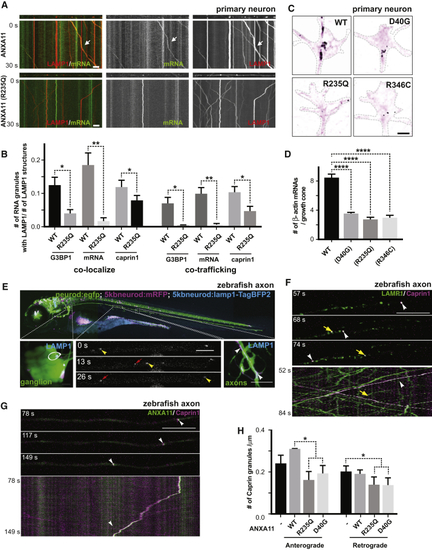
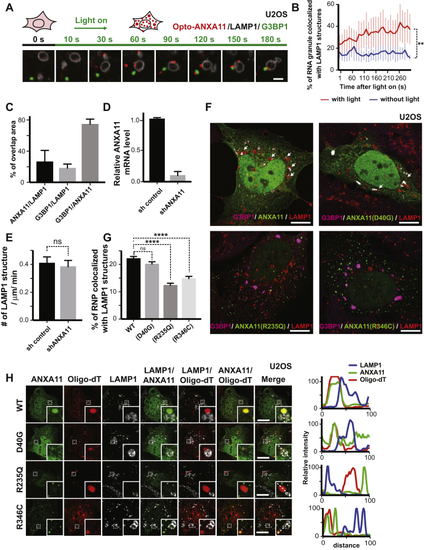
Effects of ALS-Associated ANXA11 Mutations on RNA Granule Interactions (A) U2OS cells expressing mEmerald-tagged ANXA11 (WT, D40G, R235Q or R346C) were heat shocked (43oC) for 30 minutes. A single ANXA11-positive puncta in each of the different transfected cells was photobleached and recovery of fluorescence was monitored by time-lapse imaging. Scale bar: 1 μm. (B) Quantification of the FRAP experiments in ( (C) Phase partitioning characteristics of ANXA11 ALS-associated mutants (D) U2OS cells expressing similar levels of Opto-mCherry (CRY2olig-mCherry), Opto-ANXA11 or Opto-ANXA11 ALS-associated mutant were exposed to 0.2% 488nm light to initiate oligomerization. Scale bar: 30 μm. See also (E) Quantification of integrated fluorescence intensity of Opto-labeled proteins in (F) Quantification of the number of Opto-labeled puncta present 30 minutes after the 488 nm light was turned off. n=17-19. One-way ANOVA, ns, not significant. ∗∗∗∗p < 0.0001. Error bars= SEM. (G) Immunostaining of mEmerald-tagged wild-type and mutant ANXA11 with G3BP1 and mRNA labeled by Oligo-dT in U2OS cells following 30 minutes of heat shock. Co-localization of ANXA11 with individual RNA granules is plotted in the line scans to the right. Scale bar: 30 μm. See also (H) Quantification of area of ANXA11 structures co-localizing with G3BP1-labeled RNA granules in (G). n=28-31. One-way ANOVA, ns, not significant. ∗∗∗∗p < 0.0001. Error bars = SEM. |
|
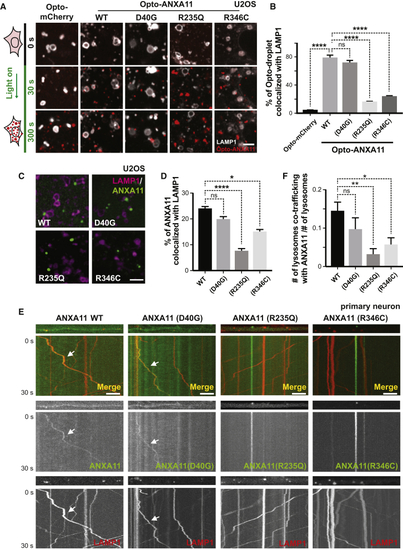
ALS-Associated Mutations in ANXA11 Disrupt Its Interactions with Lysosomes (A) Live cell imaging of Opto-mCherry, wild-type Opto-ANXA11, or mutant Opto-ANXA11 with LAMP1-HaloTag in U2OS cells before and after oligomerization induced by exposure to 488nm light. Scale bar: 2 μm. See also (B) Quantification of percentage of light-activated Opto-mcherry (CRY2olig-mCherry), wild-type Opto-ANXA11, or mutant Opto-ANXA11 clusters co-localizing with lysosomes at 300s post-488 nm light exposure from ( (C) Extent of co-localization of wild-type or mutant ANXA11 with lysosomes. U2OS cells expressing LAMP1-HaloTag, wild-type ANXA11-mEmerald or mutant ANXA11-mEmerald were imaged 30 minutes after heat shock (43oC). Scale bar: 2 μm. (D) Percentage of fluorescence associated with wild-type ANXA11 or mutant ANXA11 that co-localized with lysosomes from ( (E) Extent of co-trafficking of wild-type or mutant ANXA11 with lysosomes in axons. Axons of rat cortical neurons expressing LAMP1-HaloTag and wild-type or mutant ANXA11-mEmerald were imaged for 30 seconds. Kymographs show WT and p.D40G ANXA11 both co-traffic with lysosomes (see arrows) while p.R235Q and p.R346C each disrupt ANXA11 co-trafficking with lysosomes. Scale bar: 10 μm. (F) Number of puncta containing WT or mutant ANXA11 that co-trafficked with lysosomes as a function of total lysosome number from ( |
|
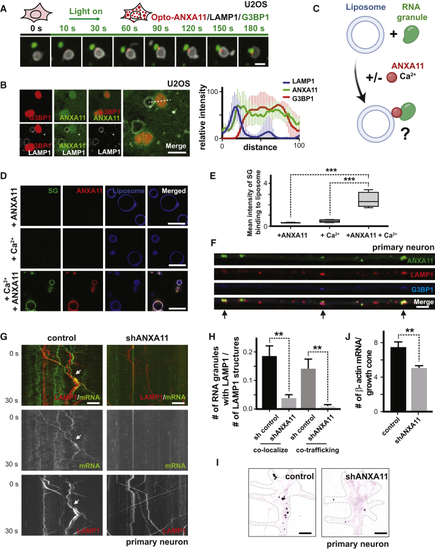
ANXA11 Acts as an Adaptor between RNA Granules and Lysosomes (A) Time-lapse imaging of U2OS cells expressing LAMP1-HaloTag, Opto-ANXA11 and mEmerald-G3BP1 after 488nm light exposure to induce Opto-ANXA11 oligomerization. U2OS cells were exposed to heat shock (43 oC) for 15 minutes prior to light activation to form visible G3BP1 stress granules. Stress granules (green) associate with LAMP1-labeled lysosomes (white) at sites where ANXA11 puncta (red) are localized. Scale bar: 1 μm. See also (B) Live cell confocal imaging of U2OS expressing LAMP1-HaloTag, ANXA11-mEmerald and mCherry-G3BP1 following 30 minutes of heat shock (43oC). Quantification of the intensity profiles of the different probes across midline of stress granules (dotted line) is shown to right. n=6, Error bars = SEM. Scale bar: 1μm. (C) Schematic of (D) Stress granule cores were purified from cultured cells, and incubated with PI3P containing liposomes +/– recombinant ANXA11 +/– Ca2+. Upper panel: + ANXA11 only, Middle panel: + Ca2+only, Bottom panel: + both ANXA11 and Ca2+. Scale bar=10 μm. (E) Quantification of mean intensity of stress granule binding to PI3P containing liposomes in (F) Co-localization of ANXA11, lysosomes, and RNA granules in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes, ANXA11-mEmerald to label ANXA11, and mCherry-G3BP1 to label RNA granules. Arrows indicate areas of ANXA11, lysosome and RNA granule co-localization. Scale bar: 5 μm. See also (G) ANXA11 knockdown perturbs mRNA/lysosome co-trafficking in axons. Kymographs of mRNA (actin-24xMBS/ MCP-NLS-2xEGFP) and lysosome (LAMP1-HaloTag) trafficking in axons is shown. Rat neurons expressed control or ANXA11-targeting shRNAs. Scale bar: 5 μm. (H) Quantification of (I) smFISH of beta-actin in growth cones from neuron expressing control shRNA (left panel) or ANXA11 shRNA (right panel). Black colored spots represent the signal from beta-actin smFISH probes, red signal represents membrane stain of growth cones. Scale bar: 1 μm. (J) Quantification of average number of beta-actin mRNA molecules in |
|
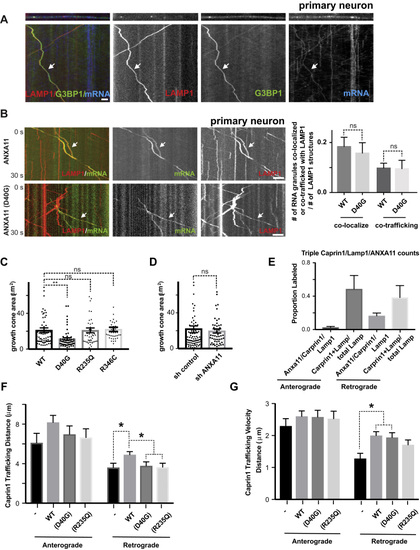
Effects of ALS-Associated ANXA11 Mutations on Axonal RNA Granule/Lysosome Hitchhiking (A) Kymographs of mRNA (actin-24xMBS/ MCP-NLS-2xEGFP ) and lysosome (LAMP1-HaloTag) trafficking in rat neuron axons expressing wild-type or R235Q mutant ANXA11. Arrows point to examples of mRNA co-trafficking with lysosomes. Scale bar: 10 μm. (B) Quantification of (C) smFISH of beta-actin in growth cones from rat neurons expressing wild-type or mutant ANXA11. Black colored spots represent the signal from beta-actin smFISH probes, red signal represents membrane stain of growth cones. Scale bar: 1 μm. (D) Quantification of (E) Lysosome trafficking in live zebrafish embryo ganglion axons. Lysosomes were labeled with LAMP1-TagBFP2 in zebrafish pLL ganglions; insets show ganglion (left) and axon tips (right). Time-lapse imaging reveals bi-directional lysosomal trafficking in these axons (bottom middle panels). (F) Imaging of live zebrafish neurons reveals bi-directional co-trafficking of CAPRIN1-positive RNA granules with lysosomes in axons. Yellow arrows point to anterograde co-trafficking of LAMP1 (green) and CAPRIN1 (magenta); white arrows point to retrograde co-trafficking of LAMP1 (green) and CAPRIN1 (magenta). Corresponding kymograph shown below. See also (G) Imaging of live zebrafish neurons expressing ANXA11 and CAPRIN1 reveals co-trafficking of ANXA11-labeled structures (green) with CAPRIN1 (magenta) in axons. Corresponding kymograph shown below. See also (H) Effect of ANXA11 ALS-associated mutations on trafficking of CAPRIN1-labeled RNA granules in zebrafish axons. CAPRIN1 and wild-type or mutant ANXA11 were expressed in zebrafish ganglion. Anterograde or retrograde trafficking of CAPRIN1 vesicles per μm along the axon length were quantified in each group. n = 9-18. Two-way ANOVA with Tukey post-hoc analysis, ∗p < 0.05. Error bars = SEM. See also |
|
RNA Granules Hitchhike on Motile Lysosomes in Mammalian Cells, Related to (A) Quantification of the percentage of RNA granules in contact with different organelles from (B) Contacting RNA granules and lysosomes are frequently in close association with ER. U20S cells were transfected with LAMP1-HaloTag, mEmerald-SEC61 and low levels of mCherry-G3BP1 for 24hrs. Cells were imaged live for 30 minutes after heat shock (43oC). Arrows point to areas where co-localized LAMP1 (white)- and G3BP1 (red)- labeled structures are in close association with Sec61-labeled ER (green). Scale bar: 1μm. (C) Quantification of velocity of G3BP1 labeled RNA granule co-localized or not co-localized with lysosomes, n= 455 (number of granules, not co-localized), 396 (number of granules, co-localized), t-test, ∗∗∗p < 0.001. Error bars = SEM. (D) Percentage of G3BP1 labeled RNA granule co-localized or not co-localized with lysosomes with displacement over 10μm, n=7 (number of cells), t-test, ∗∗p < 0.01. Error bars = SEM. (E) Percentage of G3BP1 labeled RNA granules treated or not treated with nocodazole ( (F) Time-lapse image sequence showing an RNA granule co-trafficking with a lysosome along a microtubule. U2OS cells were transfected with LAMP1-HaloTag, Ensconsin-GFP and low amounts of mCherry-G3BP1 for 24hrs. Images were acquired immediately after heat shock at 43oC. Scale bar: 1μm. (G) Quantification of LAMP1 labeled lysosomes co-localizing with G3BP1 labeled RNA granules (relative to number of lysosome), n=20 (number of cells). (H) Kymograph of mEmerald tag and lysosomes in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and PGK promoter driven mEmerald tag. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Scale bar: 5 μm. (I) Kymographs illustrating co-trafficking and stationary interaction patterns of lysosomes with RNA granules. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and actin-24xMBS/MCP-NLS-2xEGFP to label actin mRNA. Upper panel shows co-trafficking of lysosomes and mRNA, and bottom panel shows lysosomes and mRNA associating in a relatively stationary manner. Scale bar: 5 μm. (J) Kymograph of CAPRIN1-labeled RNA granules co-trafficking with lysosomes in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and mEmerald-CAPRIN1 to label RNA granules. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Arrows point to lysosomes co-trafficking with CAPRIN1-labeled structures. p50/p150Glued, doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. Scale bar: 5 μm. (K) Kymograph of TDP43-labeled RNA granules co-trafficking with lysosomes in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes and mEmerald-TDP43 to label RNA granules. Time-lapse images of axons were acquired at 100ms/frame for 30 seconds. Arrow points to a lysosome co-trafficking with a TDP43-labeled structure. Dox, doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. Scale bar: 5 μm. (L) Quantification of frequency of G3BP1, actin-MS2, CAPRIN1, TDP43 labeled RNA granule and mEmerald tag in axons, n=22(mEmerald), 19(G3BP1), 35(actin-MS2), 21(caprin1), 35(TDP43). (M) Quantification of LAMP1 labeled lysosomes co-localizing or co-trafficking with G3BP1, actin-MS2, CAPRIN1, TDP43 labeled RNA granules and mEmerald tag (relative to number of lysosomes) in axons, n=22(mEmerald), 19(G3BP1), 36(actin-MS2), 25(CAPRIN1), 41(TDP43). (N) Quantification of LAMP1-labeled lysosomes co-localizing or co-trafficking with G3BP1, actin-MS2 (relative to number of lysosomes), with or without doxycycline-inducible expression of a p50 dynactin subunit and the CC1 domain of the p150 glued subunit of dynactin. N=35(G3BP1,-), 35(G3BP1, p50/p150Glued), 36(actin-MS2, -), 30(actin-MS2, p50/p150Glued). T-test, ∗∗p < 0.01, ∗p < 0.05, ns, not significant. Error bars = SEM. |
|
Recombinant ANXA11 Undergoes Liquid-Liquid Phase Separation A. Purified ANXA11 protein formed biological condensates. Full-length wild type ANXA11 formed spherical, fusing liquid droplets at ANXA11 concentrations at 10μM facilitated by 10% dextran. Inset shows a fusion event between two phase separated liquid droplets. |
|
ANXA11 Exhibits Phase Condensate Properties and Interacts with Both RNA Granules and Lysosomes in Living Cells, Related to (A) ANXA11’s amino acid sequence 1-185 was tagged with mEmerald and expressed in U2OS cells. Small ANXA11 positive puncta appeared in cells that had not been heat shocked. (B) Live cell imaging of puncta from ( (C) Quantification of the FRAP experiment in ( (D) Co-localization of ANXA11 puncta with RNA granule markers before, during and after heat shock (HS). U2OS cells under normal culture conditions (before HS), under heat shock (HS) or 4 hours after heat shock (after HS) were fixed, hybridized with Cy3-Oligo dT(30) followed by immunostaining with antibody against TDP43. Linescans show the related intensity profiles of ANXA11 with mRNA (Cy3 Oligo-dT) and with TDP43. Scale bar: 30 μm. (E) Co-localization of ANXA11 full-length, N-terminal or C-terminal domain with RNA granules. U2OS cells were fixed after 30 minutes of heat shock (43oC), followed by immunostaining with antibody against TDP43. Line scans show the related intensity profiles of ANXA11 with TDP43. Scale bar: 30 μm. One-way ANOVA, ∗∗∗∗p < 0.0001, n=20 (FL), 11(NTD), 19(CTD), Error bars=SEM. (F) Co-localization of ANXA11 full-length, N-terminal or C-terminal domain with RNA granule. U2OS cells were fixed after 30 minutes of heat shock (43oC), followed by hybridizing with Cy3-Oligo dT(30). Linescans show the related intensity profiles of ANXA11 with TDP43. Scale bar: 30 μm. One-way ANOVA, ∗∗∗∗p < 0.0001, n=30 (FL), 30(NTD), 27(CTD), Error bars=SEM. (G) Rat cortical neurons were transduced with ANXA11-mEmerald. A single labeled ANXA11 puncta was photobleached, then the recovery of fluorescence into the bleached region-of-interest was examined over time. Scale bar: 2 μm. (H) Time-lapse imaging showing the interaction of ANXA11 puncta (red) with LAMP1-labeled lysosomes (white) in U2OS cells after heat-shock. Scale bar: 1 μm. (I) Quantification of LAMP1 labeled lysosomes co-localizing with ANXA11(relative to number of lysosomes) in U2OS or rat neuron. N=25(U2OS), 10(neuron). |
|
ALS-Associated Mutations in ANXA11 Disrupt RNA Granule Interactions, Related to (A) A schematic map of ANXA11 protein with the position of ALS-associated mutants. (B) Quantification show the temporal evolution of the integrated fluorescence intensity from the expressed Opto-mCherry, ANXA11 full-length, NTD or CTD proteins during 300 seconds of light activation, n=11 (Opto-mCherry), 17 (ANXA11 full-length), 20(ANXA11 NTD), 20(ANXA11 CTD). Error bars = SEM. (C) U2OS were transfected with Opto-mCherry (CRY2olig-mcherry), Opto-ANXA11, Opto-ANXA11 NTD or Opto-ANXA11 CTD for 24hrs. Cells with similar Opto-ANXA11 expression levels were exposed to 0.2% 488nm light to initiate oligomerization. Scale bar: 30 μm. (D) Co-localization of ANXA11 or ANXA11 ALS-associated mutants with TDP43 and mRNA labeled by Oligo-dT. U2OS cells expressing mEmerald labeled ANXA11 or ANXA11 ALS-associated mutants were heat shocked for 30 mins, fixed, and then hybridized with Cy3-Oligo dT(30) and immunostained with antibodies against TDP43 to label RNA granules. The extent of co-localization of ANXA11 or the ALS-associated mutants with the RNA granules is plotted in the line-scans to the right. (E) Quantification of percentage of area of ANXA11 structures co-localizing with TDP43-labeled RNA granules in ( (F) Co-localization of ANXA11 or ANXA11 ALS-associated mutants with RNA granules labeled by TDP43 and mRNA labeled by Oligo-dT after heat shock (HS). U2OS were heat shocked for 30 mins and then moved to 37oC for 4 hrs to allow recovery. The cells were then fixed, hybridized with Cy3-Oligo dT(30) followed by immunostaining with antibodies against TDP43 to label RNA granules. Linescan analysis show the related intensity profiles of ANXA11 or ALS-associated mutations with mRNA (Cy3 Oligo-dT) and TDP43. Scale bar: 30 μm. (G) Co-localization of ANXA11 or ALS-associated ANXA11 mutants with RNA granules labeled by G3BP1 and mRNA (right panel) after heat shock (HS). U2OS were heat shocked for 30 mins and then moved to 37oC for 4 hrs to allow recovery. The cells were then fixed, hybridized with Cy3-Oligo dT(30) followed by immunostaining with antibodies against G3BP1 (right panel) to label RNA granules. Linescan analysis show the related intensity profiles of ANXA11or ALS-associated ANXA11 mutants with mRNA (Cy3 Oligo-dT) and G3BP1. Scale bar: 30 μm. (H) U2OS cells expressing mCherry-G3BP1 to label RNA granules were co-transfected with ANXA11-mEmerald, ANXA11(D40G)-mEmerald, ANXA11(R235Q)-mEmerald or ANXA11(R346C)-mEmerald for 24 hrs. Cells were heat shocked (43oC) for 30 min, A single G3BP1-positive puncta in each of the different transfected cells was photobleached and recovery of fluorescence into the puncta was monitored by time-lapse imaging. Scale bar: 1 μm. (I) Quantification of H. N=7(WT), 9(D40G), 8(R235Q), 7(R346C). Error Bars=SEM. |
|
ALS-Associated Mutations in ANXA11 Disrupt Its Interactions with Lysosomes, Related to (A) Co-localization of light-activated opto-ANXA11 or ANXA11 N-terminal domain or C-terminal domain with lysosomes in cells. U2OS cells were co-transfected with LAMP1-HaloTag, Opto-mcherry (CRY2olig-mcherry), Opto-ANXA11, Opto-ANXA11 NTD or Opto-ANXA11 CTD for 24 hrs. Cells with similar Opto-ANXA11 expression levels were exposed to 0.2% 488nm light to initiate oligomerization. Cells were imaged over 300 seconds of light activation. Scale bar: 2 μm. (B) Percentages of light-activated Opto-mcherry (CRY2olig-mcherry), Opto-ANXA11 Opto-ANXA11 NTD or Opto-ANXA11 CTD clusters co-localizing with lysosomes after 300 seconds of light activation from the experiment in ( (C) Frequency of LAMP1 labeled vesicles in axons expressed ANXA11 or ALS-associated ANXA11 mutants. n=25(WT), 50(D40G), 15(R235Q), 22(R346C). One-way ANOVA, ns, not significant. Error bars = SEM. |
|
ANXA11 Acts as an Adaptor between RNA Granules and Lysosomes, Related to (A) Additional example of a time-lapse imaging sequence of U2OS cells expressing LAMP1-HaloTag, Opto-ANXA11 and mEmerald-G3BP1 after 0.2% 488nm light activation to initiate Opto-ANXA11 oligomerization. U2OS cells were exposed to heat shock (43 oC) for 15 minutes prior to light activation. Here, G3BP1-labeled RNA granules (green) associate with ANXA11 puncta (red) in the cytoplasm before redistributing onto the surface of LAMP1-labeled lysosomes (white) as merged puncta. Scale bar: 1 μm. (B) Quantification of the percentage of RNA granules co-localizing with LAMP1 over 300s with or without light activation in (A). N=11. Paired t-test, ∗∗, p<0.01. Error bars = SEM. (C) Percentage of co-localization between ANXA11 and LAMP1, G3BP1 and LAMP1, G3BP1 and ANXA11. N=20. Error bars = SEM. (D) Q-PCR shows relative ANXA11 mRNA level in control shRNA or ANXA11 shRNA transduced neurons. N=3. Error bars = SEM. (E) Frequency of LAMP1-labeled vesicles in axons expressed control shRNA or ANXA11 shRNA. N=36(sh control), 35 (sh ANXA11), t-test, ns, not significant. Error bars = SEM. (F) Effect of ALS-associated ANXA11 mutants on RNA granule-lysosome association in heat shocked cells. U2OS cells were transfected with mEmerald tagged ANXA11 or ANXA11 ALS-related mutant constructs. Cells were heat shocked (43oC) for 30 minutes, fixed, followed by immunostaining with antibodies against G3BP1 and LAMP1 to examine the effects of ANXA11 mutants on RNA granule-lysosome contacts compared to those in WT ANXA11-mEmerald expressing cells. Arrows point to G3BP1-labeled RNA granule(megenta) contacting with lysosomes(red). Scale bar: 30 μm. (G) Percentage of G3BP1-labeled granules co-localized with LAMP1-labeled lysosomes in ( (H) U2OS cells were transfected with mEmerald tagged ANXA11 or ANXA11 ALS-related mutant constructs. Cells were heat shocked (43oC) for 30 minutes, fixed, hybridized with Cy3-Oligo dT(30) followed by immunostaining with antibodies against LAMP1 to examine the effects of ANXA11 mutants on RNA granule-lysosome contacts compared to that in WT ANXA11-mEmerald expressing cells. Graphs on the right represent intensity profiles across the dotted line, revealing the ANXA11 mutants R235Q and R346C show decreased colocalization with RNA granules and lysosomes and affected RNA granule-lysosome contact. Scale bar: 1μm. |
|
ALS-Associated Mutations in ANXA11 Disrupt RNA Granule Hitchhiking on Lysosomes in Axons from Rat Cortical Neurons and Zebrafish Neurons, Related to (A) Kymographs showing RNA granule protein and mRNA co-transport on the same lysosomes in axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes (red), and mCherry-G3BP1 to label RNA granules (green), and actin-24xMBS/ MCP-NLS-2xEGFP(blue). Time-lapse movies of axons were then acquired at 100ms/frame for 30 seconds and displayed in kymograph format. Arrows point to a example of both G3BP1 and mRNA co-trafficking with lysosomes. Scale bar: 5 μm. (B) Kymographs showing the effect of ALS-associated ANXA11 mutants on RNA co-trafficking with lysosomes in rat neuron axons. Rat cortical neurons were transduced with LAMP1-HaloTag to label lysosomes (red), actin-24xMBS/ MCP-NLS-2xEGFP (green) and ANXA11 (upper panel) or ANXA11 (D40G) (bottom panel). Time-lapse movies of axons were then acquired at 100ms/frame for 30 seconds and displayed in kymograph format. Arrows point to an example of RNA granule co-trafficking with lysosomes. Scale bar: 5 μm. Quantification of RNA granule co-localization or co-trafficking with lysosomes in axons expressing either ANXA11 or ANXA11 p. D40G mutation (relative to lysosome number). n=36 (WT), 12 (D40G). One-way ANOVA. ns, not significant. Error bars = SEM. (C) Quantification of growth cone area in axons expressing either ANXA11 or mutant ANXA11. N=61(WT), 56(D40G), 46(R235Q), 39(R346C). One-way ANOVA. ∗∗, p<0.01, ns, not significant. Error bars = SEM. (D) Quantification of growth cone area in axons expressing either ANXA11 or mutant ANXA11. N= 80(sh control), 54(sh ANXA11). unpaired t-test. ns, not significant. Error bars = SEM. (E) Quantification of vesicles triple labeled with CAPRIN1/LAMP1/ANXA11 over the total LAMP1 vesicles undergoing anterograde or retrograde transport in zebrafish axons. N=9-18. (F) Quantification of trafficking distance for CAPRIN1 vesicles in zebrafish axons. N=9-18. Two-way ANOVA with Tukey post-hoc analysis, ns, not significant. ∗p < 0.05. Error bars = SEM. (G) Quantification of trafficking velocity for CAPRIN1 vesicles in zebrafish axons. N=9-18. Two-way ANOVA with Tukey post-hoc analysis. ∗p < 0.05. Error bars = SEM. |
Reprinted from Cell, 179, Liao, Y.C., Fernandopulle, M.S., Wang, G., Choi, H., Hao, L., Drerup, C.M., Patel, R., Qamar, S., Nixon-Abell, J., Shen, Y., Meadows, W., Vendruscolo, M., Knowles, T.P.J., Nelson, M., Czekalska, M.A., Musteikyte, G., Gachechiladze, M.A., Stephens, C.A., Pasolli, H.A., Forrest, L.R., St George-Hyslop, P., Lippincott-Schwartz, J., Ward, M.E., RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether, 147-164.e20, Copyright (2019) with permission from Elsevier. Full text @ Cell