- Title
-
Specific fibroblast subpopulations and neuronal structures provide local sources of Vegfc-processing components during zebrafish lymphangiogenesis
- Authors
- Wang, G., Muhl, L., Padberg, Y., Dupont, L., Peterson-Maduro, J., Stehling, M., le Noble, F., Colige, A., Betsholtz, C., Schulte-Merker, S., van Impel, A.
- Source
- Full text @ Nat. Commun.
|
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
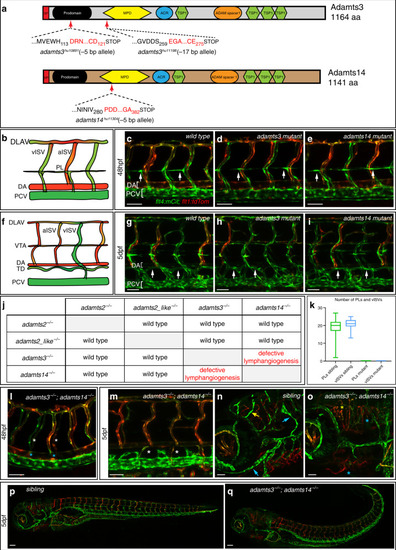
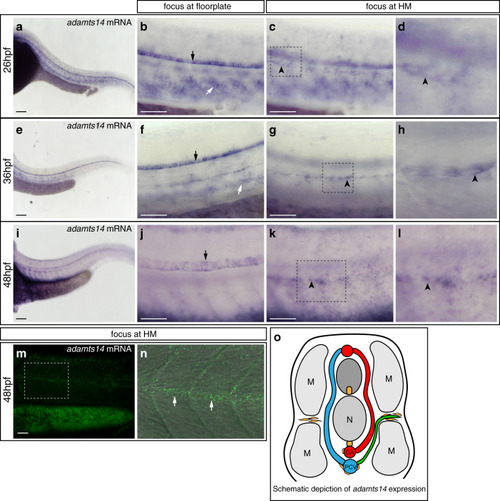
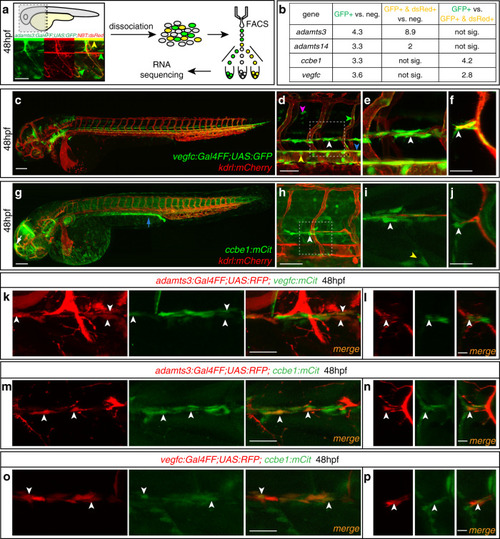
In situ hybridization against vegfc using a milder proteinaseK treatment reveals expression at the horizontal myoseptum. a, b) vegfc transcripts were detected in the hypochord, dorsal aorta (arrow) and in developing intersegmental arteries at 26hpf. b) Higher magnification of the trunk region with prominent staining of the hypochord (arrow). c, d) At 36hpf vegfc transcripts were detected in cells at the HM (black arrow), the hypochord and the dorsal aorta (orange arrow). d) Higher magnification of the trunk region. e, f) Expression of vegfc within the DA and in cells at the HM persist at 48hpf. f) High magnification of the trunk focusing at the HM level. Scale bars in a, c, e: 100μm; b, d, f: 50μm. hpf: hours post fertilization, DA: dorsal aorta, HM: horizontal myoseptum. |
|
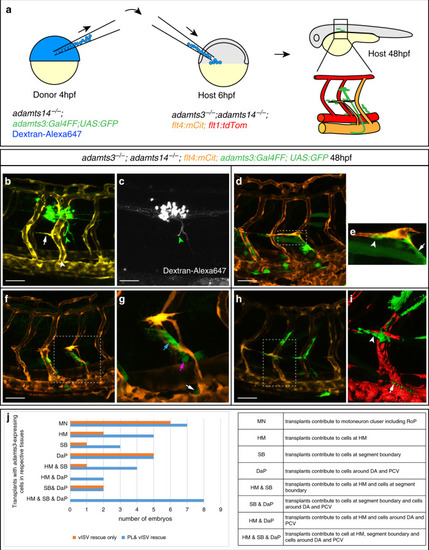
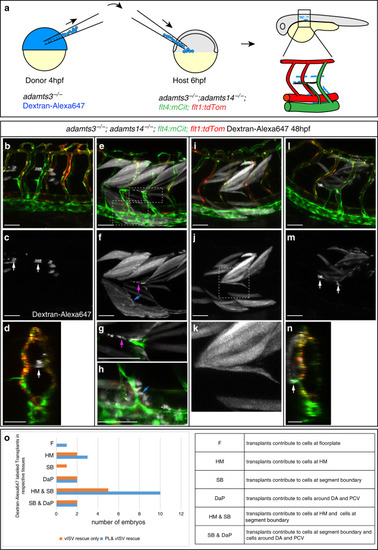
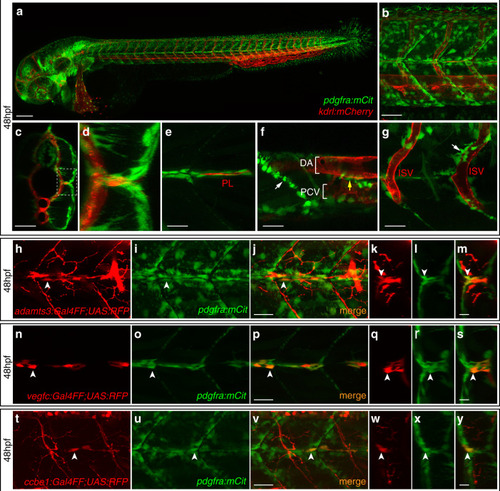
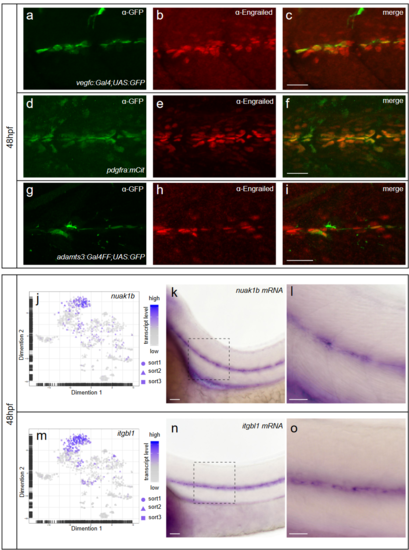
Fibroblasts at the horizontal myoseptum express Engrailed proteins. a-i) HM region of 48hpf embryos that were stained with anti-Engrailed (red) and anti-GFP (green) antibodies. a-c) All cells expressing the vegfc:Gal4FF; UAS: GFP reporter at the HM do co-express Engrailed proteins. d-f) fpdg-fra:mCitrine positive fibroblasts at the HM are also positive for Engrailed. g-i) Partial z-projections of the HM region reveal a co-expression of the adamts3 reporter and Engrailed proteins within fibroblasts. j, m) Two additional examples of genes whose transcripts are highly enriched within fibroblast cluster 2 are nuak1b and tgbl1 (see Supplementary Tables 1-3). The mRNA of both genes (k, l, n, o) can be detected specifically at the midline by ISH at 48hpf, thereby validating the notion that the cells in cluster 2 represent the fibroblast subpopulation located at the HM. Scale bars in a-i: 25 μm; k, l, n and o: 50 μm. hpf: hours post fertilization, HM: horizontal myoseptum EXPRESSION / LABELING:
|