- Title
-
Runx1 promotes scar deposition and inhibits myocardial proliferation and survival during zebrafish heart regeneration
- Authors
- Koth, J., Wang, X., Killen, A.C., Stockdale, W.T., Potts, H.G., Jefferson, A., Bonkhofer, F., Riley, P.R., Patient, R.K., Göttgens, B., Mommersteeg, M.T.M.
- Source
- Full text @ Development
|
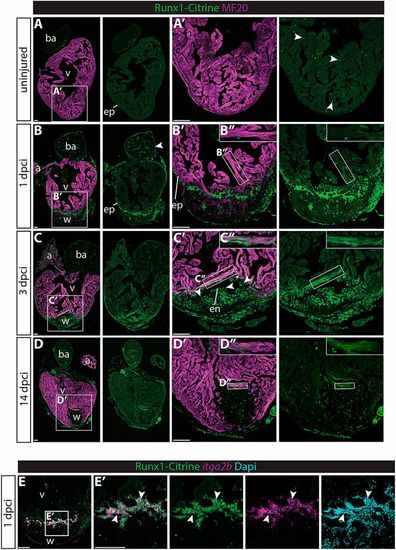
Runx1-Citrine becomes strongly expressed in the heart after cryo-injury. (A-D″) Immunohistochemistry for Runx1-Citrine (GFP antibody) and the myocardial marker MF20 at different time points after cryo-injury. (A,A′) Citrine expression in the uninjured hearts was confined to a small number of cells scattered around the heart (arrowheads). (B-B″) At 1 dpci, the epicardium was Citrine positive (arrowheads). Bright blood cells were visible within the wound and there was dim expression of Citrine overlapping with MF20 (B″). (C-C″) At 3 dpci, the epicardium, endocardium (arrowheads) and other wound cells were positive for Citrine. In addition, the myocardium in the border zone next to the wound was highly Citrine positive (C″). (D-D″) Expression of Citrine diminishes at 14 dpci but is still visible, especially in the myocardium (D″). (E,E′) In situ hybridisation for itga2b with immunohistochemistry for Runx1-Citrine and nuclear marker DAPI. Arrowheads indicate overlap of Runx1-Citrine with itg2b mRNA, indicating that thrombocytes are positive for Runx1-Citrine. a, atrium; ba, bulbus arteriosus; dpci, days post cryo-injury; en, endocardium; ep, epicardium; v, ventricle; w, wound. Scale bars: 100 μm. EXPRESSION / LABELING:
|
|
Different wound composition and faster regeneration in runx1 mutant compared with wild-type hearts. (A-E) AFOG staining of wild-type and runx1 mutant ventricles at five different time points after injury. (F,G) Quantification of the difference in wound size between the wild type and mutant at the different time points, measured by the percentage of the compact wall not yet closed (F) and the percentage of wound area compared with total ventricle area (G). n=5 per time point, two-way ANOVA with Sidak test. *P<0.05 and **P<0.01. Box extends from the 25th to 75th percentiles and whiskers indicate minimum to maximum with all data points shown. (H-K) Quantification of differences in wound composition between the fish at 7 dpci (collagen, blue; fibrin, bright red; all other cells, including myocardium/blood cells are orange), n=5. c, collagen; f, fibrin; v, ventricle; w, wound. Scale bars: 100 μm. PHENOTYPE:
|
|
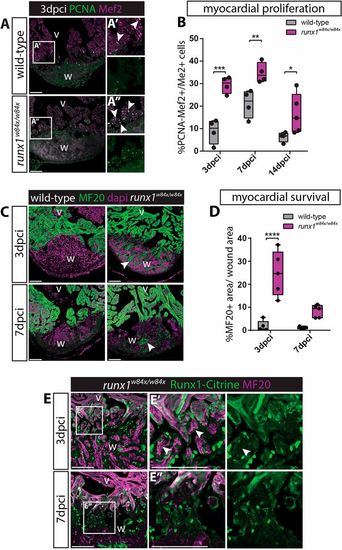
Increased myocardial proliferation and protection in the runx1 mutant. (A-A″) Immunohistochemistry for PCNA and Mef2 on 3 dpci sections. An increased number of double-positive cells (arrowheads) seems present in the mutant compared with the wild-type wound border. (B) Quantification of PCNA-positive proliferating Mef2-positive myocardial cells after injury shows increased myocardial proliferation in the runx1 mutant at all time-points analysed. n≥4, two-way ANOVA with Sidak test. (C) Immunohistochemistry for MF20 with the nuclear marker Dapi. Arrowheads indicate the presence of MF20-positive myocardial cells in the wound in the mutant at both 3 and 7 dpci. (D) Quantification of the MF20-positive area in the wound on sections in the wild type and mutant shows increased presence of myocardial cells in the mutant. n=5, two-way ANOVA with Sidak test. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. Box extends from the 25th to 75th percentiles and whiskers indicate minimum to maximum with all data points shown. (E) Immunohistochemistry for Citrine and MF20. Arrowheads indicate the surviving MF20-positive cells in the mutant wound that are Runx1-Citrine negative. v, ventricle; w, wound. Scale bars: 100 µm. |
|
Runx1-Citrine positive endocardial cells appear in the wound after injury. (A-D,F,G) Immunohistochemistry analysis for Citrine- and mCherry-positive cells in the Tg(BAC-runx1P2:Citrine;kdrl:Hsa.HRAS-mCherry) line. (A,A′) The wound at 1 dpci, with the box highlighting the flat and weakly Citrine- and mCherry-positive endocardial cells in the wound (arrowhead). (B-C′) Clearly visible and round Citrine- and mCherry-positive cells in the wound at 3 dpci (B,B′, arrowheads), but not further away from the wound (C,C′). (D) At 14 dpci, not many double-positive cells are visible anymore. (E) Quantification of the number of Citrine and mCherry double-positive cells in and away from the wound, n=4, two-way ANOVA with Tukey's test. *P<0.05, **P<0.01 and ***P<0.001. Box extends from the 25th to 75th percentiles and whiskers indicate minimum to maximum with all data points shown. (F,G) runx1 mutant wounds have a reduced number of double-positive cells (arrowheads). en, endocardium; v, ventricle; w, wound. Scale bars: 100 μm. |
|
Single cell sequencing of Citrine- and mCherry-positive cells. (A) Experimental design of selection of cells for single cell sequencing using the 10x Genomics platform. (B) UMAP plot of all cells combined, clustering into 27 different clusters. (C) Annotation of the different cell clusters. (D) UMAP plot separated into citrine/runx1-positive cells, mcherry/kdrl-positive cells and double-positive cells. (E) UMAP plot separated into wild-type uninjured cells, wild-type 3 dpci cells and runx1 mutant 3 dpci cells. Arrowheads indicate the shift in endocardial/endothelial cells, with C0 and C2 appearing, and C3 reducing in size after injury. HSC, haematopoietic stem cells; mt, mitochondrial. |
|
(A,B) All wild-type uninjured and 3 dpci Tg(BAC-runx1P2:Citrine;kdrl:Hsa.HRASmCherry) double-positive cells visualised in an UMAP plot. (A) Expression of collagen 1a1b specifically in 3 dpci double-positive cells. (B) Cell clustering within the double-positive population identifies six different cell clusters. (C) Heatmap showing that both clusters 4 and 5 express high levels of collagens, whereas cluster 4 specifically expresses smooth muscle genes. (D) myh11a and tagln are expressed in cluster 4 after injury. Arrowhead indicates myh11a expression in cell cluster 4. (E-F) Immunohistochemistry for Citrine, mCherry and Myh11. Arrowheads indicate expression of Myh11 in Tg(BAC-runx1P2:Citrine;kdrl:Hsa.HRAS-mCherry) double-positive cells at 3 (E,E′) and 7 (F) dpci. en, endocardium; v, ventricle; w, wound. Scale bars: 100 μm. |
|
(A) UMAP plot of Tg(BAC-runx1P2:Citrine;kdrl:Hsa.HRASmCherry) double-positive cells in the 3 dpci runx1 mutant as well as uninjured and 3 dpci wild-type cells. Arrowheads indicate the double-positive cluster 4 that is absent in the runx1 mutant. (B) Violin plot showing that the absent cluster 4 is the cluster most strongly expressing smooth muscle genes in the wild type after injury. (C) Immunohistochemistry for Citrine, Myh11 and MF20, showing reduced staining for Myh11 in the runx1 mutant wound at 7 dpci compared with the wild type. Arrowheads indicate overlap of Myh11 with Citrine in the endocardium. (D) Quantification of the area of Myh11 expression in the wound on sections between mutants and wild types. n=5, unpaired two-tailed t-test. *P<0.05. Box extends from the 25th to 75th percentiles and whiskers indicate minimum to maximum with all data points shown. (E,E′) In situ hybridisation for itga2b combined with immunohistochemistry for Citrine and Myh11 with nuclear marker Dapi. Arrowheads indicate Myh11-expressing blood cells that express the thrombocyte marker itga2b. (F,G) UMAP plot of all cells confirms expression of myh11a in the itga2b-positive thrombocyte cluster. v, ventricle; w, wound. Scale bars: 100 μm. |
|
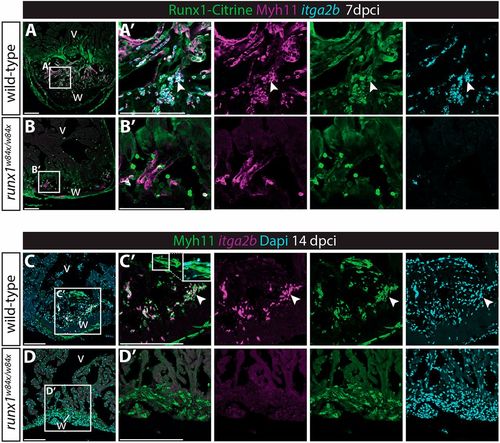
Myh11-positive endocardial cells and thrombocytes retain their double identity. (A-B′) In situ hybridisation for itga2b combined with immunohistochemistry for Citrine and Myh11. Arrowheads indicate Myh11-positive itga2b-positive thrombocytes present in the wild-type wound that are largely missing in the runx1 mutant wound at 7 dpci. (C-D′) In situ hybridisation for itga2b combined with immunohistochemistry for Myh11 with nuclear Dapi staining. Both the endocardium (inset) and thrombocytes (arrowheads) still express Myh11 in the wild-type wound at 14 dpci, while being absent in the mutant wound. v, ventricle; w, wound. Scale bars: 100 μm. |
|
Reduction in myofibroblast numbers in the runx1 mutant. (A) UMAP plot combining cluster 8 and 10 from Fig. 4B, showing very few cells in these clusters in the uninjured wild type, but the appearance of both populations after injury in the wild type and runx1 mutant. (B) UMAP plot from A, indicating expression levels of tcf21, myh11a, tagln and col1a1b. The epicardial cluster 8 expresses tcf21, whereas myofibroblast cluster 10 expresses myh11a. Both clusters express tagln and col1a1b. (C) Staining of 7 dpci sections confirms presence of Tagln and absence of Myh11 in the epicardium (arrowheads). (D) Single cell data showing the numbers of cells per cluster per sample. Increased number of epicardial cells and a reduced number of myofibroblast cells in the runx1 mutant compared with the wild type. (E) Dotplot showing expression levels of smooth muscle, EMT and collagen genes per sample in clusters 8 and 10. Increased cell numbers and expression of myofibroblast genes in both the epicardial and myofibroblast clusters in the injured wild-types compared to the uninjured wild-types. Epicardial myofibroblast gene expression is lower in the runx1 mutant compared to the uninjured wild-type, while the number of myofibroblast cells is strongly reduced in the mutant. (F,G) Immunohistochemistry for Citrine and Myh11 on 7 dpci sections. Analysis of Myh11 on 7 dpci sections confirmed the reduction in myofibroblast cell numbers close to the epicardium (arrowheads). ep, epicardium; v, ventricle; w, wound. Scale bars: 100 μm. |
|
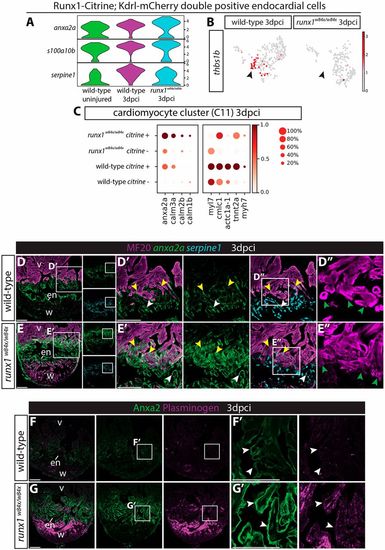
Runx1 mutant hearts upregulate Anxa2 and plasminogen. (A) Violin plots showing upregulation of anx2a and s100a10b in wild-type runx1/citrine;mcherry/kdrl double-positive cells after injury, with even higher expression in the runx1 mutant. In contrast, serpine1 is downregulated in the mutant cells. (B) UMAP plot of the runx1/citrine;mcherry/kdrl double-positive cells showing thbs1b-expressing cells. Arrowheads indicate thbs1b expression mainly in cluster 4 from Fig. 6A, which is missing in the runx1 mutant after injury. (C) Dotplot showing that anxa2a, calm1b, calm2b and calm3a are upregulated in the mutant citrine-positive myocardium at 3 dpci, whereas sarcomere genes are upregulated in the wild-type citrine-positive myocardium. (D-E′) Section in situ hybridisation for anxa2a and serpine1 with immunohistochemistry for MF20 shows that anxa2a has a similar expression pattern to Runx1-Citrine after injury in the wild type, but is expressed at much higher levels in the mutant endocardium (white arrowheads) and myocardium (yellow arrowheads). serpine1 expression is found in both the wound border endocardium (white arrowheads) and myocardium (yellow arrowheads). (D″,E″) Sarcomere structure differs between the wild-type and the mutant in the wound border (green arrowheads). (F-G′) Immunohistochemistry for plasminogen and Anxa2 shows upregulation of plasminogen in the area where Anxa2 is upregulated (arrowheads) in the mutant. en, endocardium; v, ventricle; w, wound. Scale bars: 100 μm. |