- Title
-
Copper induce zebrafish retinal developmental defects via triggering stresses and apoptosis
- Authors
- Zhao, G., Sun, H., Zhang, T., Liu, J.X.
- Source
- Full text @ Cell Commun. Signal.
|
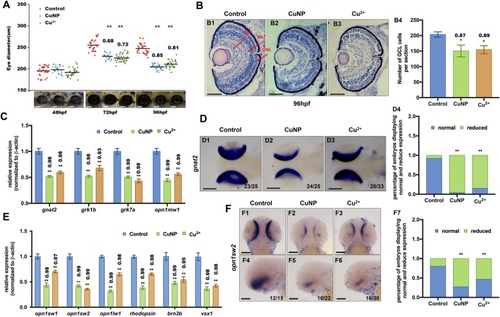
Retinal developmental defects in copper stressed embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
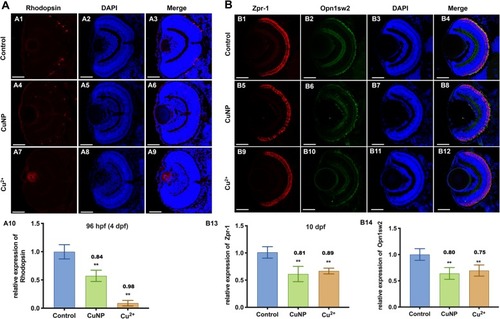
Protein levels of retinal photoreceptor in copper stressed embryos. |
|
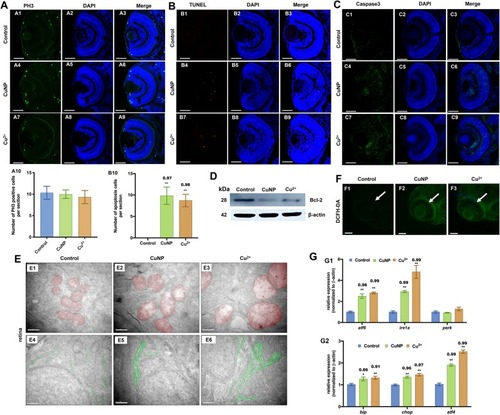
Cell proliferation, apoptosis, and oxidative & ER stresses in copper-stressed embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
ER stresses in copper-stressed and BFA-treated embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
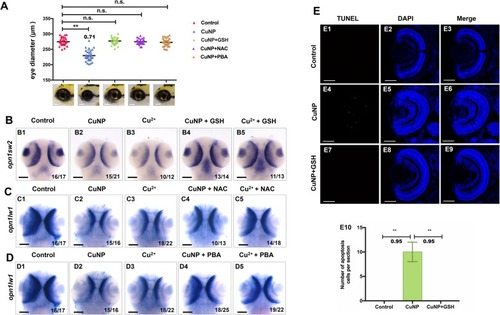
Rescue of retinal defects with the addition of ROS scavenger and ER stress scavenger. ROS scavengers GSH & NAC and ER scavenger PBA recovered the copper induced small eye defects ( |
|
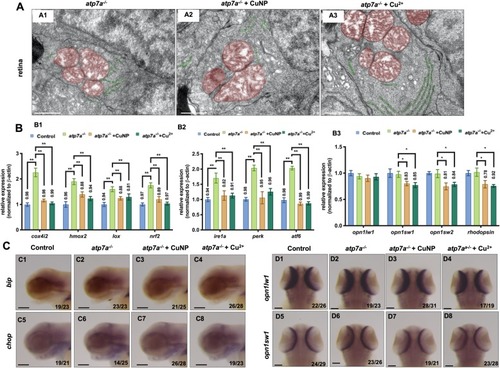
Retina development in |
|
Retina development in |
|
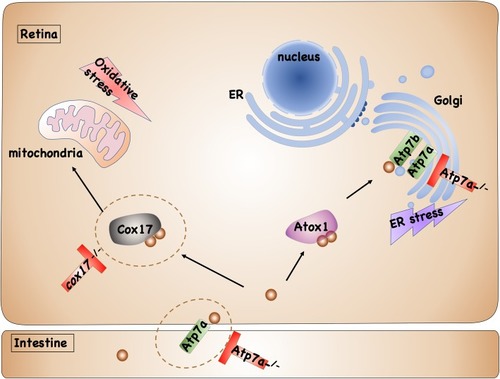
Model of copper induced retinal developmental defects in WT, |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |