- Title
-
Function of Arl4aa in the Initiation of Hematopoiesis in Zebrafish by Maintaining Golgi Complex Integrity in Hemogenic Endothelium
- Authors
- Guo, Y., Cheng, B.Y.L., Wang, D., Ma, A.C.H., He, B.L., Man, T.K., Cheung, M.P.L., Shi, X., Ng, N.K.L., Leung, A.Y.H.
- Source
- Full text @ Stem Cell Reports
|
Expression Pattern of arl4aa in Zebrafish Embryos and Adults (A–C) Whole-mount in situ hybridization (WISH) of arl4aa at (A) 18 and 24 hpf, (B) 36 hpf, and (C) 48 hpf. (D and E) Cross-sections of the trunk at 24 (D) and 36 hpf (E) as shown by H&E staining (left panels) and WISH (right panels), at the level shown by the black lines in (A and B). H&E, hematoxylin and eosin; hpf, hours postfertilization; NC, notochord; DA, dorsal aorta; PCV, posterior cardinal vein. (F) Expression of arl4aa in different adult zebrafish tissues detected by semi-quantitative RT-PCR. Scale bars, 250 μm (A–C) and 50 μm (D and E). EXPRESSION / LABELING:
|
|
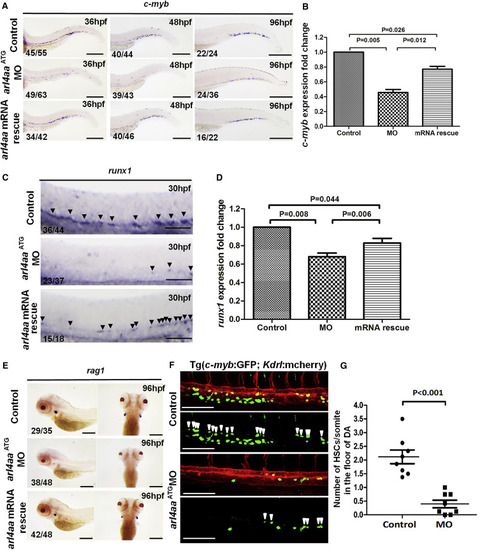
Knockdown of Zebrafish arl4aa Perturbed Definitive Hematopoiesis (A) c-myb expression in un-injected control (top), arl4aa morphant (MO) (middle), and morphant rescued by arl4aa mRNA (bottom) at 36 (left), 48 (middle), and 96 hpf (right). (B) qRT-PCR of c-myb expression in control, MO, and arl4aa mRNA rescue groups (mRNA rescue) in embryos at 48 hpf. Data are representative of three independent experiments with error bars representing the mean ± SEM. (C) runx1 expression at 30 hpf in un-injected control (top), arl4aa ( middle), and morphant rescued by arl4aa mRNA (bottom). Arrowheads indicate runx1-expressing cells in the ventral wall of the dorsal aorta (DA). (D) qRT-PCR of runx1 expression in control, MO, and arl4aa mRNA rescue groups (mRNA rescue) in embryos at 30 hpf. Data are representative of three independent experiments with error bars representing the mean ± SEM. (E) rag1 expression at 96 hpf between control (top), MO (middle), and arl4aa mRNA rescue groups (bottom). (F) Confocal microscope imaging of double transgenic (cmyb:GFP; kdrl:mCherry) embryos injected with arl4aa MO. Arrowheads denote cmyb + kdrl + HSCs along the DA. The embryos were examined at 36 hpf. (G) Number of HSCs at the floor of the DA per somite in Tg (cmyb:GFP; kdrl:mcherry) embryos at 36 hpf. The results are presented as mean ± SEM (p < 0.001; n = 8 embryos per group in three independent experiments). Scale bars, 250 μm (A) and 100 μm (C and E). See also Figure S2. |
|
Knockout of Zebrafish arl4aa Perturbed Definitive HSC Development in F0 Somatic Mutants (A) Knockout of arl4aa by a TALEN pair (E2-2) targeting the translation start site. Upper panel: the TALEN pair; middle panel: genotyping of single embryos (numbers 1–5) injected with TALEN E2-2. Mean knockout efficiency was 51% ± 5.5% (n = 5); lower panel: DNA sequence of three mutant clones from F0 embryos. (B) Knockout of arl4aa by two TALEN pairs targeting exons 1 and 2 (E1 + E2). Upper panel: the TALEN pairs, primer positions indicated by red arrows. Middle panel (top): genotyping of single embryos (numbers 1–4) injected with E1 + E2, showing an amplicon of 319 bp. Middle panel (bottom): knockout efficiency was based on qPCR amplifying arl4aa intron 1 existing fold, which was deleted by E1 + E2. Mean knockout efficiency was 26.3% ± 2% (n = 10). Knockout efficiency was correlated with hematopoietic phenotype, being 31% ± 2.6% (n = 5) and 21.6% ± 0.7% (n = 5) for embryos whose c-myb expression was completely absent and reduced. Lower panel: DNA sequencing of three mutant clones from F0 embryos. (C) c-myb expression in control (left arm TALEN E2-2L) (upper), E2-2 injected (middle), and E2-2 and arl4aa mRNA co-injected (lower) embryos at 48 hpf. (D) c-myb expression in control (top), E1 + E2 injected embryos showing absent (second) and reduced (third) c-myb expression, and E1 + E2 and arl4aa mRNA co-injected (bottom) embryos. Data are shown as means ± SEM. Scale bars, 250 μm (in WISH). The numbers in each WISH showed the numbers of embryos with typical gene expression as shown over the total number of embryos injected in each group. |
|
Knockout of Zebrafish arl4aa Perturbed Definitive Hematopoiesis in F2 Mutants (A) Strategy of raising F2 mutant embryos. (B) DNA sequencing and genotyping of F2 mutant embryos containing 2-bp deletion and 6-bp substitutions. The amino acids are labeled under the DNA sequence. (C) Expression of c-myb in F2 embryos (upper, WT; middle, heterozygous; lower, homozygous) at 36 (left panel) and 48 hpf (middle panel) and of rag1 at 96 hpf (right panel). (D and E) Intensity of c-myb expression in CHT at 48 hpf (D) and rag1 expression in thymus at 96 hpf (E) as measured by ImageJ software. Data are shown by means ± SEM; n and p values are indicated in (D) and (E). Scale bars, 250 μm (in WISH). The numbers in each WISH showed the numbers of embryos with typical gene expression as shown over the total number of embryos injected in each group. |
|
Deletion of arl4aa and ARL4A Caused Fragmentation of Golgi Complex in VDA Endothelial Cells in Zebrafish and Human HeLa Cells (A) Electron microscopy of endothelial cells of VDA of un-injected (control [CON]) and F0 embryos injected with TALEN E2-2 at 30 hpf. (i–ii) Endothelial cells of VDA are shown in the rectangles. (iii–vi) High power magnification of control (iii) and F0 embryos (iv–vi). Golgi complex shows typical configuration in control embryos (9/10 cells from 4 control embryos) but aberrant morphology in F0 embryos being unusually elongated with disruption (iv, 6/10 cells from 6 embryos), under-developed (v, 1/10 cells from 6 embryos), and “onion-skin” appearance (vi, 2/10 cells from 6 embryos). NC, notochord; VDA, ventral wall of dorsal aorta; PCV, posterior cardinal vein; N, nucleus. (B) Electron microscopy of neural tube in CON (i) and F0 embryos (ii) injected with TALEN E2-2, showing normal Golgi complex configuration (iii and iv). Scale bars, 2 μm (Ai,ii and Bi,ii) and 200 nm (Aiii–vi and Biii, iv). Orange arrowheads: abnormal morphology of the Golgi complex. (C) Immunostaining of Golgi membrane Giantin in control and arl4aa morphant (MO) embryos at 30 hpf. The insert on top shows the morphology of 30-hpf embryos with the rectangle highlighting the region of the DA under detailed examination. The dotted red line in the bright field defines the DA. Immunostaining: Giantin was stained with anti-Giantin followed by Alexa Fluor 488-conjugated antibody and the nuclear outline is shown by DAPI staining. The yellow arrowhead denotes the fragmentation of the Golgi complex in endothelial cells from the VDA. The white arrowhead denotes normal expression of Giantin at the Golgi complex. Scale bar, 100 μm. (D) Immunostaining of Giantin followed by Alexa Fluor 594-conjugated antibody in scramble sequence control, ARL4A shRNA-1, and ARL4A shRNA-2 transfected HeLa cells. GFP+ labeled the successfully transfected cells. White arrowheads indicate normal Golgi complex and yellow arrowheads denote the fragmented and dispersed Giantin staining. Scale bar, 20 μm. (E) Both ARL4A shRNA-1 and shRNA-2 significantly increased the proportion of cells with fragmented Golgi complex as shown by Giantin expression, data are shown as means ± SEM (n = 3 experiments, p < 0.001). See also Figures S4–S6. |
|
Knockdown of arl4aa Blocked Cleavage of Notch Receptor Precursor and Downregulated Notch Signaling Pathway along DA Floor (A) Expression of Notch signaling genes in control and arl4aa morphant groups at 42 hpf (notch1, notch3, hey2, and her5) and 33hpf (gata2a and gata2b). Data are representative of three independent experiments with error bars representing the mean ± SEM. (B) Western blot showing Notch1 and NICD protein expression in control, arl4aa morphant embryos, and DAPT-treated embryos at 42 hpf. The numbers indicate the image intensities of immunoblotting relative to those of tubulin in each embryo group. The intensity in the controls was set arbitrarily as 1. The results are representative of at least three experiments. DAPT, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester. (C) Confocal microscope imaging of double transgenic tp1:eGFP; kdrl:mCherry embryos injected with arl4aa MO and examined at 30 hpf. Arrowheads indicate cells in the floor of the DA with active Notch signaling. Scale bar, 100 μm. (D) Enumeration of tp1 + kdrl + HSCs in the floor of DA (n = 8 embryos per group, p < 0.001). Data are representative of three independent experiments with error bars representing the mean ± SEM. (E) hsp70l:Gal4; UAS:NICD1a embryos injected with arl4aa MO were heat shocked at 14–15 somites and c-myb expression was performed at 36 hpf. Sibling embryos without NICD1a were similarly heat shocked as control. Scale bar, 250 μm. The number of embryos with typical c-myb expression as shown over the total number of embryos examined is shown in each panel. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|