- Title
-
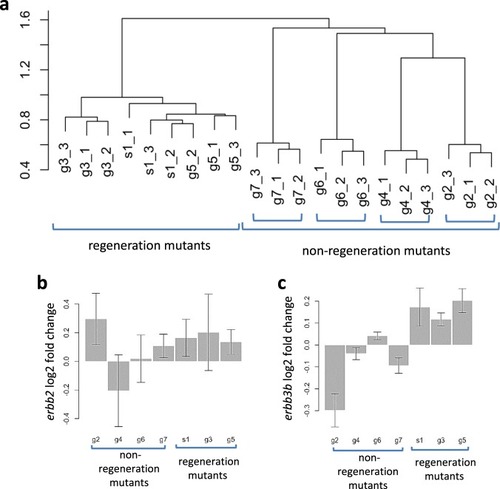
A subset of SMN complex members have a specific role in tissue regeneration via ERBB pathway-mediated proliferation
- Authors
- Pei, W., Xu, L., Chen, Z., Slevin, C.C., Pettie, K.P., Wincovitch, S., NISC Comparative Sequencing Program, Burgess, S.M.
- Source
- Full text @ NPJ Regen Med
|
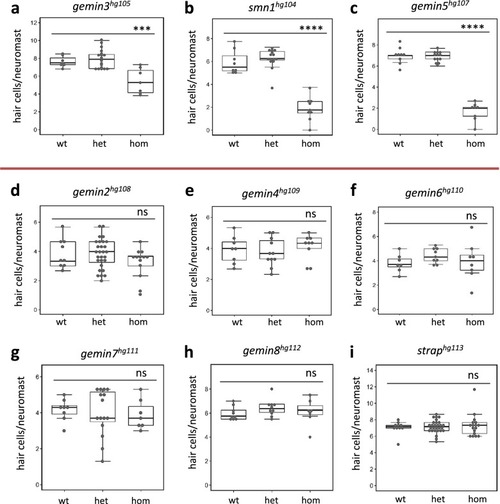
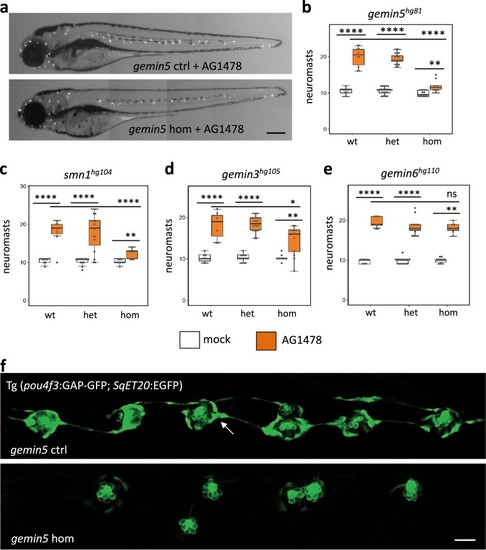
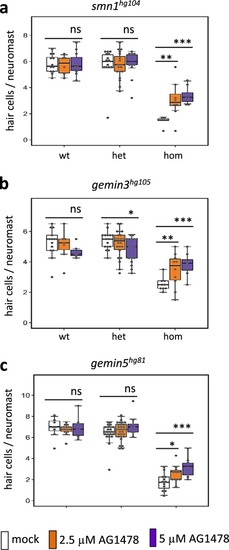
Hair cell regeneration is impaired by homozygous mutations of PHENOTYPE:
|
|
|
|
PHENOTYPE:
|
|
|
|
|
|
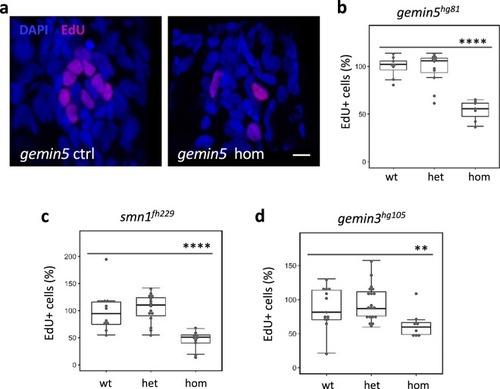
PHENOTYPE:
|
|
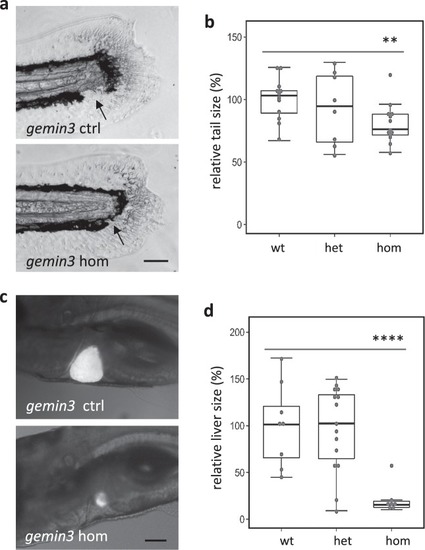
PHENOTYPE:
|
|
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|