- Title
-
Expression of tert Prevents ALT in Zebrafish Brain Tumors
- Authors
- Idilli, A.I., Cusanelli, E., Pagani, F., Berardinelli, F., Bernabé, M., Cayuela, M.L., Poliani, P.L., Mione, M.C.
- Source
- Full text @ Front Cell Dev Biol
|
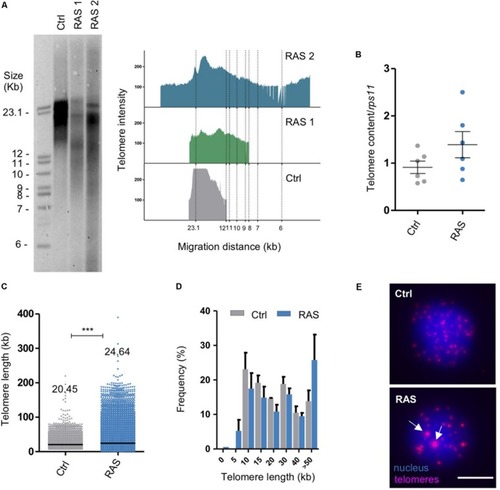
Zebrafish brain tumors have heterogeneous telomeres. PHENOTYPE:
|
|
Telomerase is not involved in telomere maintenance in zebrafish brain tumors. EXPRESSION / LABELING:
PHENOTYPE:
|
|
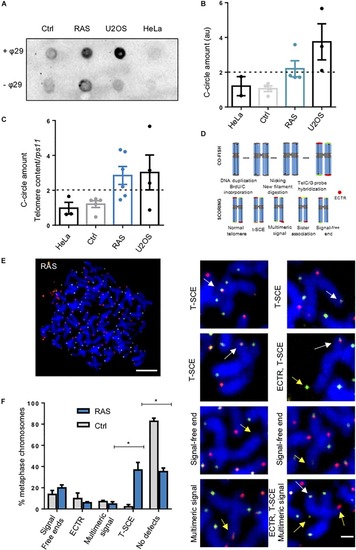
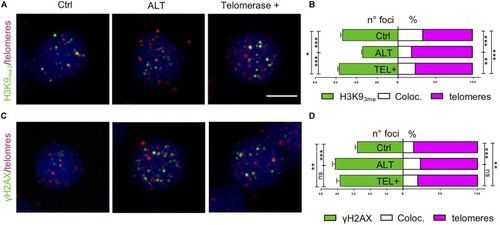
Zebrafish brain tumors are ALT. PHENOTYPE:
|
|
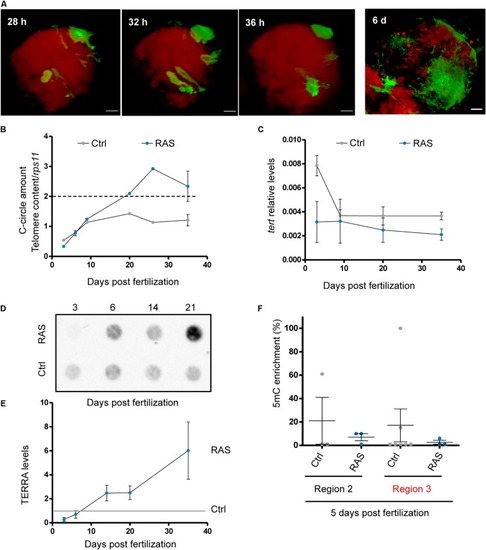
Zebrafish brain tumors exhibit elevated TERRA expression. PHENOTYPE:
|
|
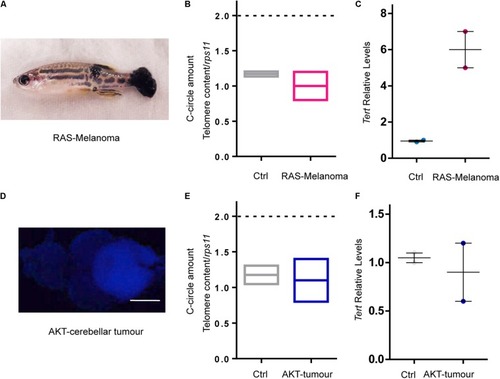
RAS expression is not the main driver of ALT in zebrafish cancer. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Development of ALT is preceded by a reduction of EXPRESSION / LABELING:
PHENOTYPE:
|
|
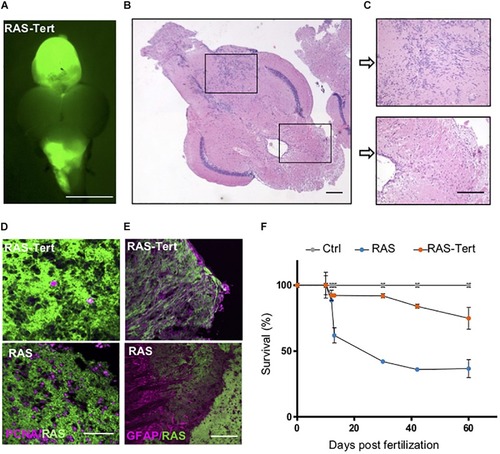
Expression of functional |
|
Expression of |
|
Expression of |