- Title
-
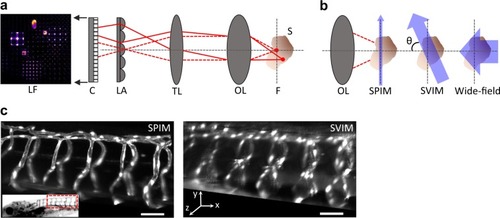
High-contrast, synchronous volumetric imaging with selective volume illumination microscopy
- Authors
- Truong, T.V., Holland, D.B., Madaan, S., Andreev, A., Keomanee-Dizon, K., Troll, J.V., Koo, D.E.S., McFall-Ngai, M.J., Fraser, S.E.
- Source
- Full text @ Commun Biol
|
|
|
|
|
|
|
Functional imaging of a 5 dpf larval zebrafish with pan-neuronal fluorescent calcium indicators, |