- Title
-
Coronary Revascularization During Heart Regeneration Is Regulated by Epicardial and Endocardial Cues and Forms a Scaffold for Cardiomyocyte Repopulation
- Authors
- Marín-Juez, R., El-Sammak, H., Helker, C.S.M., Kamezaki, A., Mullapuli, S.T., Bibli, S.I., Foglia, M.J., Fleming, I., Poss, K.D., Stainier, D.Y.R.
- Source
- Full text @ Dev. Cell
|
Regenerating Coronaries Sprout Superficially and toward the Cardiac Lumen (A) cEC proliferation at different times after injury (n = 4 ventricles per time point). (B) Ventricle at 7 dpci stained for coronaries (green), CMs (red), and DNA (blue); (Bi, Bii) high-magnification images of coronary vessels sprouting into the ventricle (white arrowheads) and superficially (magenta arrowheads). (C and D) 3D image reconstructions of the injured area after tissue clearing and staining for coronaries (red) and CMs (green). (C) Luminal view of intra-ventricular coronary sprouting (arrowheads) in the border zone (white arrows). Sprouts (yellow arrowheads) shown in high magnification (n = 3). (D) Intra-ventricular sprouting vessels in the apex (arrowheads). High-magnification images of sprouts (yellow arrowheads) shown in i and ii (n = 3). Asterisk marks the border zone. (E) Tg(-0.8flt1:RFP); TgBAC(apln:EGFP) ventricle at 96 hpci. Yellow dotted line delineates the area with apln:EGFP+ coronaries (n = 6). (F) Ventricular section at 96 hpci stained for coronaries (red), apln:EGFP expression (green), PCNA (white), and DNA (blue). High-magnification images of coronary vessels sprouting intra-ventricularly (Fi, white arrowheads) and superficially (Fii, white arrowheads) (n = 6). (G) qPCR analysis of ppargc1a, apln, ndufb5, and atp5j mRNA levels in sorted -0.8flt1:RFP+ cECs at 96 h post-sham (hps) (n = 4) and 96 hpci (n = 4). (H) qPCR analysis of ppargc1a, apln, ndufb5, and atp5j mRNA levels in sorted -0.8flt1:RFP+ cECs from 8 months post-fertilization (mpf) untouched WT (n = 4) and ppargc1a−/− (n = 4) ventricles. (I) qPCR analysis of ppargc1a, apln, ndufb5, and atp5j mRNA levels in sorted -0.8flt1:RFP+ cECs from 8 months post-fertilization (mpf) WT (n = 4) and ppargc1a−/− (n = 4) ventricles at 96 hpci. (J) cEC proliferation in 8 mpf WT (n = 5), ppargc1a−/− (n = 5), and apln−/− (n = 5) ventricles. (K) Section of a TgBAC(apln:CreERT2); Tg(ubb:GSR) ventricle at 7 dpci stained for GFP (white), mCherry (green), CMs (red), and DNA (blue); right panel shows high-magnification image of recombined coronaries (white arrows) (n = 6). White dotted lines delineate injured area. Data in graphs expressed as mean ± SEM. ns, no significant difference, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars: 50 μm. See also Figures S1–S3; Videos S1 and S2. |
|
Epicardial Chemokine Expression Directs Superficial Coronary Revascularization (A–D) Ventricles at 10 dpci stained for epicardium (white), endothelium (green), CMs (red), and DNA (blue). High-magnification images of intra-ventricular sprouting coronaries (Ai, Bi; EGFP+ yellow arrowheads) and superficial coronaries (Aii, Bii; EGFP+, magenta arrowheads) in DMSO- (A) and MTZ- (B) treated animals. Number of superficial cECs (C) and intra-ventricular coronaries (D) in DMSO- (n = 6) and MTZ- (n = 6) treated animals at 10 dpci. (E) 7 dpci ventricle stained for cxcl12b:YFP expression (white), coronary ECs (green), CMs (red), and DNA (blue). High-magnification images of regenerating epicardium upregulating cxcl12b:YFP expression. (F) qPCR analysis of aldh1a2 and cxcl12b mRNA levels in sorted tcf21:RFP+ cells from 7 days post-sham (dps) and 7 dpci ventricles (n = 4). (G–K) 7 dpci ventricle stained for cxcl12b:YFP expression (green), activated epicardium and endocardium (white), CMs (red), and DNA (blue) (G). High-magnification images of regenerating cxcl12b:YFP+ epicardium (yellow arrowheads) and activated endocardium negative for cxcl12b:YFP expression (magenta arrowheads). Wholemount images of Tg(fli1a:EGFP) WT (H) and cxcr4a−/− (I) ventricles at 7 dpci. (I) High-magnification image shows an intra-ventricular sprouting coronary in a cxcr4a−/− heart (white arrowhead). Number of superficial cECs (J) and intra-ventricular coronaries (K) in WT (n = 6) and cxcr4a−/− (n = 6) animals at 7 dpci. Data in graphs expressed as mean ± SEM. ∗∗p < 0.01. Scale bars: 100 μm. See also Figures S4 and S5; Table S1. |
|
Epicardial Hypoxia Stimulates Epicardial cxcl12b Expression and Coronary EC Proliferation via Hif1a (A) qPCR analysis of aldh1a2 and egln3 mRNA levels in sorted tcf21:RFP+ cells from 7 dps and 7 dpci ventricles (n = 4). (B and C) 7 dps (B) and 7 dpci (C) ventricles stained for epicardium (green), hypoxyprobe (red), and DNA (blue). Insets show high-magnification images of sham (B) and regenerating (C) epicardium. (D) qPCR analysis of cxcl12b mRNA levels in WT ventricles at 7 dps (n = 4) and 7 dpci (n = 4) and in hif1a−/− ventricles at 7 dpci (n = 4). (E and F) WT (E) and hif1a−/− (F) ventricles stained for endothelial nuclei (red), PCNA (green), and DNA (blue). Insets show high-magnification images of proliferating cECs (white arrowheads). (G–I) cEC proliferation in WT (n = 4), and hif1a−/− (n = 4) ventricles at 96 hpci (G). Ventricles from PBS- (H) and DMOG- (I) treated animals stained for endothelial nuclei (red), PCNA (green), and DNA (blue). Insets show high-magnification images of proliferating cECs (white arrowheads). (J) cEC proliferation in PBS- (n = 4), and DMOG- (n = 4) treated ventricles at 96 hpci. White dotted lines delineate the injured area. Data in graphs expressed as mean ± SEM. ns, no significant difference, ∗p < 0.05, ∗∗p < 0.01. Scale bars: 100 μm. |
|
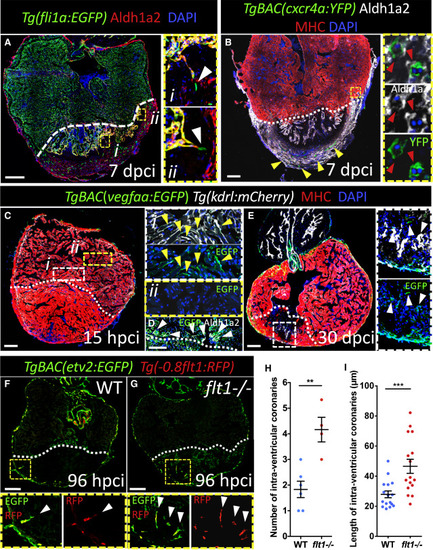
Vegfaa Signaling Directs Intra-ventricular Coronary Sprouting (A) 7 dpci ventricle stained for endothelium (green), Aldh1a2 (red), and DNA (blue). High-magnification images show intra-ventricular sprouting coronaries (EGFP+, Aldh1a2−) (white arrowhead) in close proximity to activated endocardium (EGFP+, Aldh1a2+) (n = 4). (B) 7 dpci ventricle stained for regenerating coronaries (green), Aldh1a2 (white), CMs (red), and DNA (blue). Regenerating superficial coronaries (yellow arrowheads). High-magnification images show a cross section of regenerating intra-ventricular coronaries surrounded by activated endocardium (n = 6) (red arrowheads). (C–E) 15 hpci (C and D) (n = 6) and 30 dpci (E) (n = 5) ventricles stained for vegfaa:EGFP expression (green), endocardium (white), CMs (red), activated endocardium ([D], white), and DNA (blue). High-magnification images of vegfaa:EGFP+ border zone endocardium (Ci; yellow arrowheads), endocardium distal to the injury (Cii), and vegfaa:EGFP+ activated endocardium (D and E; white arrowheads). (F and G) WT (F) and flt1−/− (G) ventricles stained for cardiac endothelium (green) and cECs (red). High-magnification images show intra-ventricular sprouting coronaries (white arrowheads). (H and I) Number (H) and length (I) of intra-ventricular coronaries in WT (n = 6) and flt1−/− (n = 4) animals at 96 hpci. White dotted lines delineate injured area. Data in graphs expressed as mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars: 100 μm. See also Figure S5. |
|
Coronaries Constitute a Scaffold Available to Cardiomyocytes during Regeneration as well as during Development (A) 7 dpci ventricle stained for coronaries (green), CMs (red), and DNA (blue) (n = 8). CMs associated with regenerating coronaries in the injured area (white arrowhead). (B) 30 dpci ventricle stained for cxcr4a:YFP expression (green), activated endocardium and epicardium (white), CMs (red), and DNA (blue) (n = 6). White arrowheads point to regenerating coronaries scaffolding the regenerating myocardium. High-magnification images of regenerating intra-ventricular coronaries scaffolding the regenerating myocardium (red arrowheads). (C) 7 dpci ventricle stained for coronaries (green), gata4:GFP expression (white), DNA (blue), and CMs (red) (n = 6). High-magnification image shows regenerating gata4:GFP+ CMs associated with coronaries in the injured area (red arrowheads). (D) 7 dpci ventricle stained for gata4:GFP expression (white), CM nuclei (red), PCNA (green), and DNA (blue). High-magnification image shows cortically located and proliferating gata4:GFP+ CMs. (E) Proliferation index of gata4:GFP+ versus gata4:GFP− CMs at 7 dpci (n = 6). (F–F”) wholemount images of hearts from 20 mm long 7 wpf fish (n = 6). High-magnification images of developing coronaries (RFP+) and cortical CMs (GFP+). White arrowheads point to vessels at the sprouting front of the developing coronary network (not associated with developing cortical CMs); yellow arrowhead points to cortical CMs (associated with coronaries). (G-G”) Ventricle from a 7 wpf fish stained for coronary ECs (white), gata4:GFP expression (red), CMs (green), and DNA (blue). Yellow arrowheads point to developing cortical CMs. Coronary vessel (white arrowhead) flanked by gata4:GFP+ CMs. White dotted lines delineate injured area. Data in graph expressed as mean ± SEM. ∗∗∗p < 0.001. Scale bars: 100 μm (A–F); 50 μm (G). See also Figure S6. |
|
Perturbations in Coronary Revascularization Impair Cardiomyocyte Repopulation (A and B) WT (A) and cxcr4a−/− (B) ventricles at 7 dpci stained for CMs (red) and DNA (blue). Presence (A) and absence (B) of cortically located regenerating CMs is indicated by white arrowhead and asterisk, respectively. (C–G) Distance spanned into the injured area by cortically located CMs in WT (n = 6) and cxcr4a−/− (n = 6) at 7 dpci. Control ([D], +EtOH) and recombined ([E], +4-OHT) ventricles at 90 dpci stained for CMs (red) and DNA (blue). Asterisk marks an area devoid of CMs (E). Control ([F], +EtOH) and recombined ([G],+4-OHT) ventricles at 90 dpci stained with AFOG to identify muscle (orange), collagen (blue), and fibrin (red). Asterisks mark contact areas between spared and regenerated myocardium. Magenta arrowheads point to regenerating CMs inside the remaining scar (F). Green dashed lines outline the scar area. (H–J) Quantification of scar area in control (n = 5) and recombined (n = 4) ventricles at 90 dpci (H). Ventricles from control ([I], PBS) and poly(I:C) (J) injected medaka at 14 dpci stained for CMs (red), DNA (blue), and endothelium (green). Yellow arrowheads point to CMs expanding beyond the border zone endocardium into the injured area; white arrowheads point to ECs in close proximity to CMs in the injured area. (K) Quantification of EC to CM association index in control (n = 4) and poly(I:C) (n = 4) injected medaka at 14 dpci. White (A, B, D, E, I, and J) and black (F and G) dotted lines delineate injured area. Data in graphs expressed as mean ± SEM. ∗p < 0.05, ∗∗∗∗p < 0.0001. Scale bars: 100 μm. |
Reprinted from Developmental Cell, 51, Marín-Juez, R., El-Sammak, H., Helker, C.S.M., Kamezaki, A., Mullapuli, S.T., Bibli, S.I., Foglia, M.J., Fleming, I., Poss, K.D., Stainier, D.Y.R., Coronary Revascularization During Heart Regeneration Is Regulated by Epicardial and Endocardial Cues and Forms a Scaffold for Cardiomyocyte Repopulation, 503-515.e4, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell