- Title
-
The Grass Carp Genomic Visualization Database (GCGVD): an informational platform for genome biology of grass carp
- Authors
- Tang, M., Lu, Y., Xiong, Z., Chen, M., Qin, Y.
- Source
- Full text @ Int. J. Biol. Sci.
|
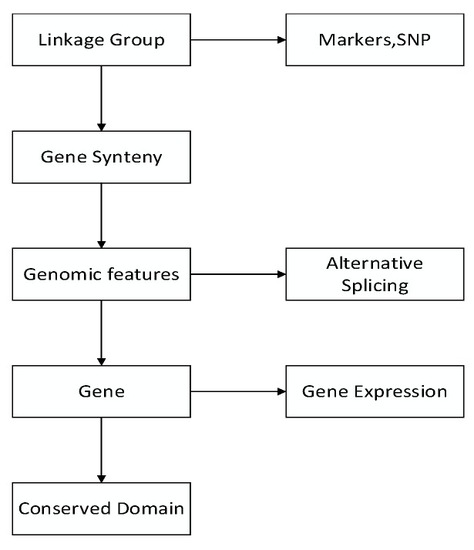
The pipeline of visualizing the genetic and genomic data of the grass carp. |
|
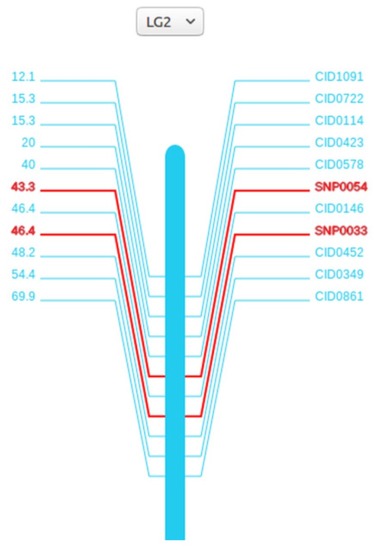
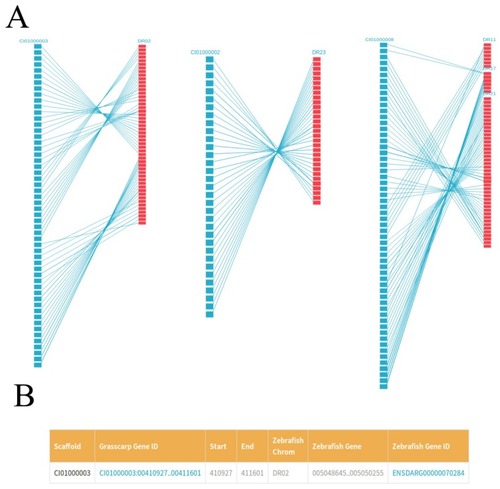
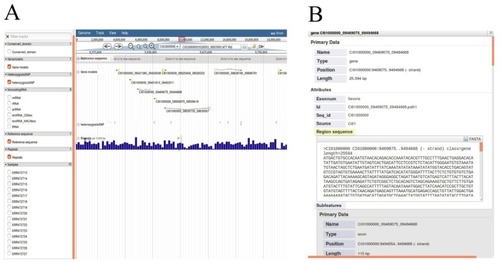
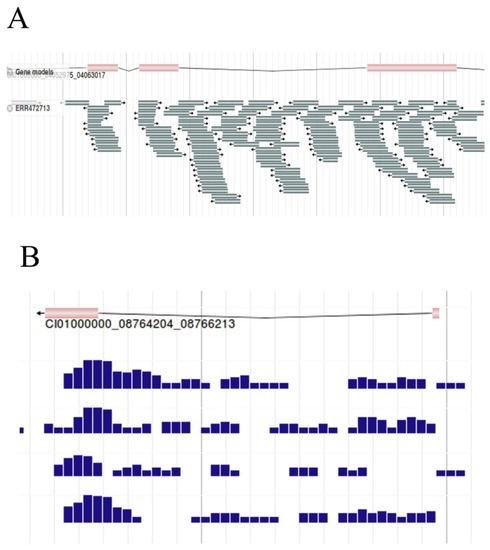
The grass carp genome annotation data model. |
|
|
|
|
|
|
|
|
|
|
|
|