- Title
-
A rare mutation in SMAD9 associated with high bone mass identifies the SMAD-dependent BMP signalling pathway as a potential anabolic target for osteoporosis
- Authors
- Gregson, C.L., Bergen, D., Leo, P., Sessions, R.B., Wheeler, L., Hartley, A., Youlten, S., Croucher, P.I., McInerney-Leo, A.M., Fraser, W., Tang, J.C.Y., Anderson, L., Marshall, M., Sergot, L., Paternoster, L., Davey-Smith, G., AOGC Consortium, Brown, M.A., Hammond, C., Kemp, J.P., Tobias, J.H., Duncan, E.L.
- Source
- Full text @ J. Bone Miner. Res.
|
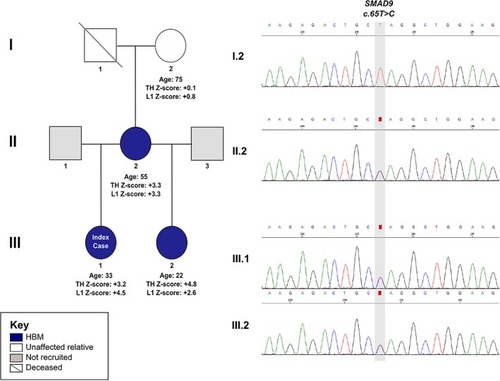
The HBM pedigree and electrophoretogram images of a segregating |
|
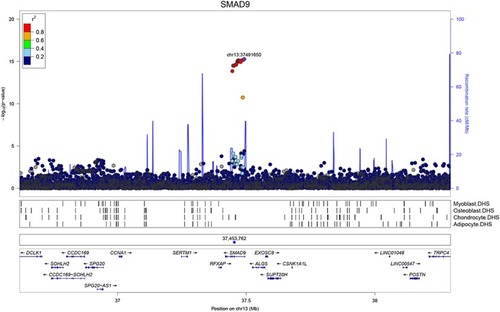
GWAS for eBMD measured in UK Biobank: Regional association plot for the locus containing |
|
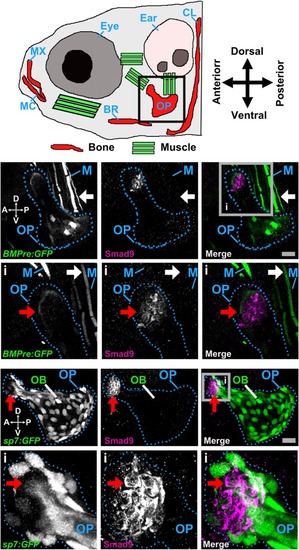
Smad9 protein expression in the larval zebrafish opercle bone. ( |
|
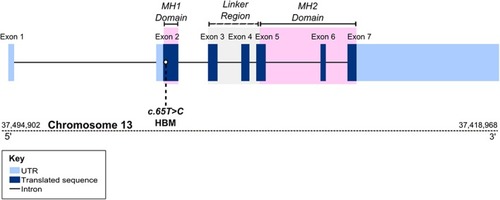
Position of c.65T>C p.Leu22Pro variant within |
|
( |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |