- Title
-
Dysregulation of Microglial Function Contributes to Neuronal Impairment in Mcoln1a-Deficient Zebrafish
- Authors
- Jin, W., Dai, Y., Li, F., Zhu, L., Huang, Z., Liu, W., Li, J., Zhang, M., Du, J., Zhang, W., Wen, Z.
- Source
- Full text @ iScience
|
Aberrant Morphology of Embryonic Microglia in biluo Mutants (A) Neutral red staining of 4-dpf siblings and mutants. Abundant microglia (white arrows) are positive for neutral red in the siblings (left) but not in the mutants. Quantification data of neutral red positive microglia number in 4-dpf siblings and mutants. n(sibling) = 7 embryos, n(biluo) = 6 embryos. ***p < 0.001 Error bars represent mean ± SD. (B) Lymphocyte cytosolic lplastin1 (Lcp1) antibody staining and DIC images of the brains of 4 dpf embryos indicate that microglia are present in biluo mutants but exhibit an abnormal enlarged morphology with accumulation of vacuoles. Quantification data of Lcp1 positive microglia number in 4 dpf siblings and mutants. White dashed lines indicate the optic tectum, yellow lines indicate the microglia presented in high magnification view on the right, and red dashed lines indicate the mutant microglia under DIC view. n(sibling)= 4 embryos, n(biluo)=6 embryos. Error bars represent mean ± SD. Aberrant Morphology of Embryonic Microglia in biluoMutants (A) Neutral red staining of 4-dpf siblings and mutants. Abundant microglia (white arrows) are positive for neutral red in the siblings (left) but not in the mutants. Quantification data of neutral red positive microglia number in 4-dpf siblings and mutants. n(sibling) = 7 embryos, n(biluo) = 6 embryos. ***p < 0.001 Error bars represent mean ± SD. (B) Lymphocyte cytosolic lplastin1 (Lcp1) antibody staining and DIC images of the brains of 4 dpf embryos indicate that microglia are present in biluo mutants but exhibit an abnormal enlarged morphology with accumulation of vacuoles. Quantification data of Lcp1 positive microglia number in 4 dpf siblings and mutants. White dashed lines indicate the optic tectum, yellow lines indicate the microglia presented in high magnification view on the right, and red dashed lines indicate the mutant microglia under DIC view. n(sibling)= 4 embryos, n(biluo)=6 embryos. Error bars represent mean ± SD. |
|
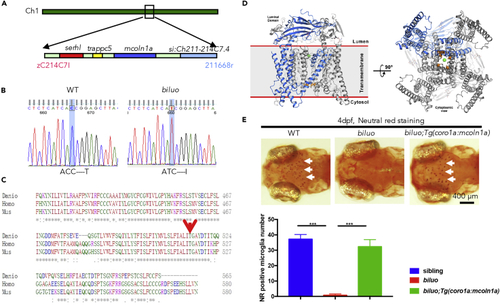
The Microglia Defect in biluo Mutants Is Caused by a Point Mutation in mcoln1a (A) Positional cloning maps the biluo mutation to an 82-kb region on chromosome 1 between two SSLP markers, zC214C7I and 211668r. This region contains four genes: serhl, trappc5, mcoln1a, and si:Ch211-214C7.4. (B) Sequencing the coding regions of the four candidate genesrevealed a C to T point mutation in the mcoln1a gene, which leads to the change of amino acid T519 to (I). (C) Alignment of human (Homo) MCOLN1, mouse (Mus) MCOLN1, and zebrafish (Danio) Mcoln1a protein sequence. Red arrow indicates the conserved 519th Threonine. (D) 3D structural model of Mcoln1a channel. Green dot represents the central pore, and orange dots indicate the amino acid T519, which is mutated to I in biluo mutants. (E) Neutral red (NR) staining of 4-dpf Tg(coro1a:mcoln1a);biluoembryos indicates that the neutral red staining defect of microglia in biluo mutants can be rescued by ectopically expressing WT mcoln1a in microglia. White arrows represent microglia. Quantification data of neutral-red-positive microglia number in 4-dpf siblings, biluo mutants, and biluo;Tg(coro1a:mcoln1a) embryos. n(sibling) = 5 embryos, n(biluo) = 5 embryos, n(biluo;Tg(coro1a:mcoln1a)) = 5 embryos. ***p < 0.001, ANOVA. Error bars represent mean ± SD. |
|
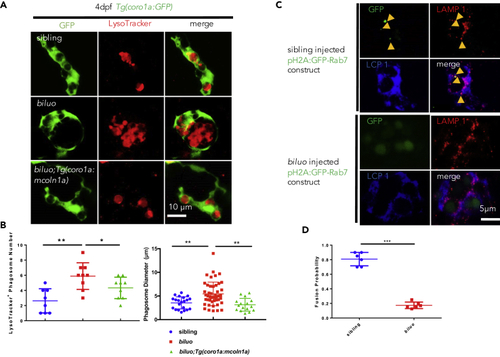
The Late Endosome and Lysosome Fusion Is Impaired in the biluo Microglia (A) Fluorescent imaging (green) and LysoTracker staining of 4-dpf sibling, biluo, and Tg(coro1a:mcoln1a);biluo embryos show that the accumulation of acid phagosomes in biluo mutant microglia (middle panel) and morphological defect of microglia in biluo mutants can be rescued by ectopically expressing WT mcoln1a in microglia (lower panel). (B) Quantification of the number and diameter of LysoTracker-positive phagosomes in the microglia in 4-dpf Tg(coro1a:GFP) siblings (blue dots), Tg(coro1a:GFP);biluo mutants (red squares), and Tg(coro1a:mcoln1a);biluo embryos (green triangles). n(sibling) = 8 microglia from 5 embryos; n(biluo) = 9 microglia from 5 embryos; n(Tg(coro1a:mcoln1a);biluo) = 9 microglia from 5 embryos. **p < 0.01, *p < 0.05, ANOVA. Error bars represent mean ± SD. (C) Anti-GFP (green), anti-LAMP1, and anti-Lcp1 (blue) triple antibody staining of WT embryos injected H2A:GFP-Rab7. Upper panels represent WT microglia (blue); late endosomes (green) are co-localized with the lysosomes. Lower panels represent mutant microglia (blue); late endosomes (green) are not co-localized with lysosomes. (D) Quantification of fusion probability of late endosomes and lysosomes in sibling microglia (blue dots) and mutant microglia (red squares). n(sibling) = 6 microglia from 4 embryos, n(biluo) = 6 microglia from 4 embryos. ***p < 0.001. Error bars represent mean ± SD. |
|
The T519 to I Mutation Reduces Mcoln1a-Mediated Ca2+ Efflux (A) Representative calcium efflux of WT (blue) and mutant Mcoln1a channel. Stars represent calcium peaks we quantified. (B) Quantification of the mean of the peak of ΔF/F0 (representing Trpml1a-mediated Ca2+efflux) in the macrophages in WT embryos injected with WT CMV:GCaMP6s-mcoln1a construct (blue dots) and the macrophages in biluo mutants injected with mutant CMV:GCaMP6s-mcoln1aT519I (red triangles). ΔF/F0 is calculated as (F-F0)/F0, where F0 is the baseline fluorescence signal. Error bars represent mean ± SD. n(WT late endosome) = 20 from 8 macrophages, n(biluo late endosome) = 20 form 7 macrophages. PHENOTYPE:
|
|
Optic Tectal Neurons in biluo Mutants Exhibit Excessive Spontaneous and Visual-Evoked Activities (A) Distribution of frequency of spontaneous responses of the tectal neurons in sibling, biluo, biluo;Tg(coro1a:mcoln1a), biluo;Tg(elavl3:mcoln1a), and biluo;Tg(coro1a:mcoln1a;elavl3:mcoln1a). n(sibling) = 214 cells in 4 embryos, n(biluo) = 166 cells in 4 embryos, n(biluo;Tg(coro1a:mcoln1)) = 165 cells in 4 embryos, n(biluo;Tg(elavl3:mcoln1a)) = 221 cells in 5 embryos and n(biluo;Tg(coro1a:mcoln1a;elavl3:mcoln1a)) = 225 cells in 4 embryos. P(sibling vs biluo) = 4.6 × 10−13, P(biluo vs biluo;Tg(coro1a:mcoln1a)) = 9.4 × 10−6, P(biluo vs biluo;Tg(elavl3:mcoln1a)) = 0.0017, and P(biluo vs biluo;Tg(coro1a:mcoln1a;elavl3:mcoln1a)) = 5.7 × 10−10. (B) Schematic of recording visual-evoked responses of optic tectal neurons in 6-dpf embryos. Red light flashed was given to the left eye of the embryos at 1, 2, 3, 4, and 5 min, and the duration of each stimulus was 2 s. The neuronal responses of the right half tectum were recorded. (C) Distribution of average peak of ΔF/F0 in sibling, biluo, biluo;Tg(coro1a:mcoln1a), biluo;Tg(elavl3:mcoln1a), and biluo;Tg(coro1a:mcoln1;elavl3:mcoln1). n(sibling) = 203 cells in 4 embryos, n(biluo) = 263 cells in 4 embryos, n (biluo;Tg(coro1a:mcoln1a)) = 305 cells in 4 embryos, n(biluo;Tg(elavl3:mcoln1a)) = 364 cells in 5 embryos, n(biluo;Tg(coro1a:mcoln1a;elavl3:mcoln1a)) = 280 cells in 4 embryos. P(sibling vs biluo) = 4.9 × 10−17, P(biluo vs biluo;Tg(coro1a:mcoln1a)) = 4.7 × 10−22, P(biluo vs biluo;Tg(elavl3:mcoln1a)) = 4.2 × 10−6, P(biluo vs biluo;Tg(coro1a:mcoln1a;elavl3:mcoln1a)) = 3.2 × 10−34, and P(biluo;Tg(coro1a:mcoln1a) vs biluo;Tg(elavl3:mcoln1a)) = 2.1 × 10−11. (D) Representative images of neuron (green)-microglia contact in 5-dpf sibling, biluo, biluo;Tg(coro1a:mcoln1a), and biluo;Tg(elavl3:mcoln1a) embryos. (E) Quantification of contact probability between microglia and optic tectal neurons. Contact duration longer than 24 s is recognized as a functional microglia-neuron contact. n(sibling) = 9 embryos, n(biluo) = 10 embryos, n (biluo;Tg(coro1a:mcoln1a)) = 9 embryos, n (biluo;Tg(elavl3:mcoln1a)) = 8 embryos. ***p < 0.0001, **p < 0.001, ANOVA. Error bars represent mean ± SD. |