- Title
-
Cxcr1 mediates recruitment of neutrophils and supports proliferation of tumor-initiating astrocytes in vivo
- Authors
- Powell, D., Lou, M., Barros Becker, F., Huttenlocher, A.
- Source
- Full text @ Sci. Rep.
|
Expression of oncogenic Kras from an astrocyte-specific promoter drives transformation in the larval hindbrain. Live-imaging of mosaic gfap:GFP, gfap:GFP-krasWT, and gfap:GFP-krasG12V expression at 3 days post-fertilization (dpf) in the larval zebrafish hindbrain (A). Red dashed outline in schematic of A shows approximate field of view and image orientation for all figures. White dashed lines in representative images denote the hindbrain region of interest in all figures. Morphology of expressing cells was categorized based on characteristics observed throughout the population of injected larvae and larvae were assigned to category of most severe cell morphology identified (B, n = 19 gfap:GFP, 17 gfap:krasWT, and 38 gfap:krasG12V larvae; Chi-squared analysis performed). gfap:H2B-mCherry was mosaically co-expressed with gfap:kras plasmids to label nuclei of Kras-expressing cells (C). qRT-PCR was performed on whole-larvae isolates for EMT genes including vimentin, mmp9, cdh1 (e-cadherin), and cdh2 (n-cadherin). Cq were normalized to housekeeping gene rps11 and then normalized to krasWT condition (D, n = 5 krasWT and 5 krasG12V replicates). Larvae were fixed at 3 dpf or 6 dpf and immunostained for N-cadherin (E). N-cadherin co-localization was observed in gfap:krasG12V-expressing masses at 6 dpf (yellow asterisks). Kras-expressing larvae were fixed at 3 dpf and immunostained for proliferation marker phospho-Histone H3 (pH3, F). pH3/GFP double positive cells were quantified using confocal optical sectioning (single optical slice from yellow dashed box shown in inset) and normalized to the total gfap-expressing area (G, n = 16 larvae/condition). *p < 0.05. |
|
Neutrophils are recruited to the tumor-initiating microenvironment. Live-imaging of neutrophils in the hindbrain of Tg(mpx:mCherry) larvae expressing gfap:krasWT or krasG12V at 3 dpf (A). Neutrophils within the field of view were quantified and normalized to the total gfap-expressing area within the same field of view at 3 and 6 dpf (B, n = 24 krasWT 3dpf, 42 krasG12V 3 dpf, 24 krasWT 6 dpf, and 30 krasG12V 6 dpf larvae). Live time-lapse imaging was performed for 9 hours beginning at 3–3.5 dpf and total neutrophils recruited to the hindbrain during the period of imaging were quantified (C, n = 9 krasWT and 11 krasG12V larvae). qRT-PCR was performed on whole-larvae mRNA isolates for several neutrophil recruiting chemokines implicated in the tumor microenvironment including cxcl8a, cxcl8b, and il1b (D, n = 6 replicates for cxcl8a/b, 5 replicates for il1b, normalized as in Fig. 1D). *p < 0.05. EXPRESSION / LABELING:
|
|
Blocking neutrophil motility reduces proliferation of tumor-initiating cells. Wildtype or Tg(mpx:mCherry) embryos were co-injected with gfap:GFP-krasG12V plasmid and rac2 or standard control morpholino (MO). Larvae were live-imaged for neutrophil recruitment at 3 dpf (A) and quantified for neutrophils within the field of view normalized to GFP-expression area (B, n = 13 control MO and 18 rac2 MO larvae). Larvae were also fixed at 3 dpf and immunostained for phospho-Histone H3 (C). Double-positive (pH3 and GFP) cells were quantified by optical sectioning and normalized as above (D, yellow arrowheads in C, n = 21 control MO and 17 rac2 MO larvae). Tg(mpx:mCherry-2a-Rac2WT) or Tg(mpx:mCherry-2a-Rac2D57N) embryos were injected with gfap:GFP-krasG12V and live-imaged at 3 dpf (E). mCherry-positive neutrophils were quantified and normalized as above (F, n = 52 Rac2WT and 46 Rac2D57N larvae). Larvae were fixed at 3 dpf and immunostained for phospho-Histone H3 (G). pH3-GFP double positive cells were quantified by optical section and normalized as above (H, n = 30 Rac2WT and 37 Rac2D57N larvae). *p < 0.05. EXPRESSION / LABELING:
|
|
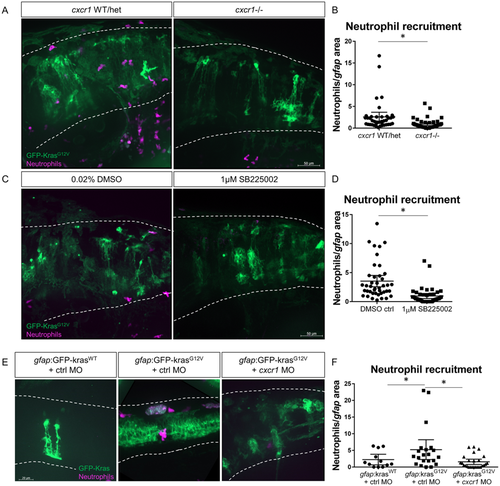
Cxcr1 promotes neutrophil recruitment to the tumor-initiating microenvironment. Adult cxcr1 heterozygote zebrafish expressing Tg(mpx:mCherry) were in-crossed, injected with gfap:krasG12V, imaged for neutrophil recruitment to the hindbrain at 3 dpf, and then processed for single-embryo genotyping (A). Neutrophils recruited to the field of view were quantified in cxcr1 mutants and WT or heterozygote siblings (no difference observed between +/+ and +/−) and normalized to total gfap-expressing area (B, n = 40 WT/het and 44 cxcr1 mutant larvae). Tg(mpx:mCherry) larvae expressing gfap:krasWT or krasG12V were treated beginning at 2.5 dpf with 1 µM SB225002 or 0.02% DMSO vehicle control for 16 hours followed by live-imaging (C). Neutrophil recruitment was quantified and normalized as above (D, n = 42 DMSO and 43 SB225002 larvae). Wildtype Tg(mpx:mCherry) embryos were co-injected with gfap:GFP-krasWT or krasG12V and standard control MO or cxcr1 splice-blocking morpholino. Larvae were live-imaged at 3 dpf (E) and neutrophil recruitment was analyzed as described previously (F, n = 12 krasWT, 21 krasG12V ctrl MO, and 26 krasG12V cxcr1 MO larvae). *p < 0.05, One-way ANOVA with post-hoc Tukey’s test performed for data in 4 F. |
|
Blocking neutrophil chemotaxis signaling reduces proliferation of tumor-initiating cells. cxcr1 heterozygote fish were in-crossed, injected with gfap:krasG12V, fixed for pH3 immunostaining at 3 dpf, and genotyped post-imaging as described above (A). pH3-GFP double-positive cells were quantified and normalized as in previous figures (B, yellow arrowheads in A, n = 19 WT/het and 26 cxcr1−/− larvae). Wildtype larvae expressing gfap:krasG12V were treated with 1 µM SB225002 or DMSO control as described in Fig. 4. Larvae were fixed at 3 dpf and immunostained for phospho-Histone H3 (C). Proliferating cells were quantified as above (D, double-positive cells marked by yellow arrowhead in C, n = 17 larvae/condition). *p < 0.05. |
|
Inhibition of neutrophil chemotaxis reduces early mass formation in p53 mutant larvae. Morphology of gfap:GFP-krasG12V expressing astrocytes in WT or p53 mutant background at 3 and 6 dpf (A, n = 87 WT 3dpf, 113 p53−/− 3dpf, 75 WT 6 dpf, and 99 p53−/− 6dpf larvae; Chi-squared analysis performed). The percentage of larvae developing masses at 6 dpf is shown in (B). p53 mutant embryos were injected with gfap:GFP-krasG12V plasmid and treated with DMSO control or 1uM SB225002 beginning at 2.5 dpf with drug refreshed every 24 hrs. Larval hindbrain was live-imaged at 3 dpf and 6 dpf (6 dpf shown in (C), asterisks denote masses). Morphology of GFP-expressing cells was categorized as described in Fig. 1B (D, n = 84 DMSO 3 dpf, 88 SB225002 3 dpf, 39 DMSO 6 dpf, and 50 SB225002 6 dpf larvae; Chi-squared analysis performed). Average number of masses was quantified in all larvae at 6 dpf (E, n = 40 DMSO and 50 SB225005 larvae). Larvae were also categorized by number of masses formed (0,1, or ≥ 2) by 6 dpf (F) and in larvae with masses present, average mass size (μm2) was measured (G, n = 29 DMSO and 15 SB225002 larvae). *p < 0.05 For Supplementary Figure Legends, please see Supplementary Information file. |
|
Cxcr2 is dispensable for neutrophil recruitment to tumor-initiating astrocytes. Adult cxcr2 heterozygote zebrafish expressing Tg(mpx:mCherry) were in-crossed, injected with gfap:krasG12V, imaged for neutrophil recruitment to the hindbrain at 3 dpf, and then processed for single-embryo genotyping (A). Neutrophils recruited to the field of view were quantified in cxcr2 mutants and WT or heterozygote siblings (no difference observed between +/+ and +/-) and normalized to total gfap-expressing area (B, n= 24 WT, 23 cxcr2+/-, 7 cxcr2-/-). Data pooled from 2 replicates. |
|
Cxcr1/2 inhibition with neutrophil-specific motility loss. Tg(mpx:mCherry-2a-Rac2WT) or Tg(mpx:mCherry-2a-Rac2D57N) embryos were injected with gfap:GFP-krasG12V and treated with 1μM SB225002 or DMSO control as described in Figure 4. Larvae were fixed at 3 dpf and immunostained for phospho-Histone H3 (A). pH3-GFP double positive cells were quantified by optical section and normalized as above (B, n= 10 Rac2WT + DMSO, 19 Rac2D57N + DMSO, 17 Rac2D57N + SB225002). Data pooled from 2 replicates, *p<0.05. |