- Title
-
Divergent Hemogen genes of teleosts and mammals share conserved roles in erythropoiesis: analysis using transgenic and mutant zebrafish.
- Authors
- Peters, M.J., Parker, S.K., Grim, J., Allard, C.A.H., Levin, J., Detrich, H.W.
- Source
- Full text @ Biol. Open
|
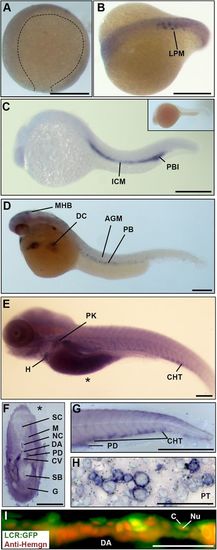
Hemogen expression in zebrafish embryos. (A-H) Wild-type embryos, WISH. (A) Epiboly at 9 hpf. Hemogen expression was not detected. (B) Ten-somite stage. Hemogen transcripts along the lateral plate mesoderm (LPM). (C) 20 hpf. Hemogen staining in the intermediate cells mass (ICM) and posterior blood island (PBI). The inset shows a sense probe control. (D) 33 hpf. Hemogen-positive primitive erythrocytes of the peripheral blood (PB) exited the Ducts of Cuvier (DC) onto the yolk. Staining at the midbrain-hindbrain boundary (MHB) was observed. (E) 144 hpf. Hemogen expression in the caudal hematopoietic tissue (CHT) and pronephric kidney (PK) and in erythrocytes in the heart (H). The asterisk indicates the plane of the cross-section in panel F. (F) 144 hpf. Cross-section of embryo in panel E showing heavily stained pronephric ducts. (G) 48 hpf. Lateral aspect of tail. Hemogen transcripts in the CHT and pronephric tubule duct (PD). (H) Kidney touch print from adult fish. Hemogen expression was observed in proerythroblasts (ProE) and normoblasts (N) but not in erythrocytes (E). (I) 48 hpf. View of circulating EGFP+ erythrocytes in the dorsal aorta (DA) of Tg(Lcr:EGFP)cz3325Tg zebrafish after staining for Hemogen protein by indirect immunofluorescence. Hemogen (red signal) accumulated in nuclei (Nu) of erythrocytes whereas the cytoplasm (C) was marked by EGFP. Other abbreviations: AGM, aorta gonad mesonephros; PT, pronephric tubule; CV, caudal vein; DA, dorsal aorta; G, gut; M, myotomes; NC, notochord; SB, swim bladder; SC, spinal cord. Scale bars: (A-F) 250 µm; (G) 1 mm; (H) 100 µm; (I) 50 µm. EXPRESSION / LABELING:
|
|
Alternative promoters drive Hemogen expression in hematopoietic and nonhematopoietic tissues in zebrafish. WISH of wild-type embryos (A-B) and adult tissues (C-H). (A) 48 hpf. Hemogen expression in the pronephric kidney glomeruli (PG), pronephric tubule duct (PD), caudal hematopoietic tissue (CHT) and brain (Br). (B) 48 hpf. Section showing strong Hemogen expression in the hindbrain (HB) but at low levels in the midbrain (MB). (C,D) Dorsal (C) and ventral (D) views of the adult zebrafish brain after staining for Hemogen transcripts. CC, crista cerebellaris; Hy, hypothalamus; EG, eminentia granularis. (E) Schematic drawing of the dorsal view. Hemogen was highly expressed at the midbrain-hindbrain boundary within the EG, in the CC and in the Hy. The asterisk indicates the plane of the cross-section in panel F. (F) Section of the hindbrain showing Hemogen expression in the periventricular gray zone (PGZ). (G) Hemogen was expressed by Sertoli cells (SE) between the seminiferous tubules (ST) of the testes. (H) Hemogen was expressed in early (I-III) but not late (IV) stage oocytes. Transcripts accumulated around the germinal vesicle (GV). (I) Schematic of the Hemogen noncoding exons 1T and 1H (gray) upstream of the first coding exon (white); bent arrows, transcription initiation sites. Arrowheads mark primer binding sites for qPCR amplification of transcripts initiated from exons 1T or 1H. (J) Expression of transcripts from alternative promoters determined by qRT-PCR using RNA from blood, testes and ovaries of adult TU zebrafish. Expression in three biological replicates were normalized to β-actin and calculated relative to ovaries. Error bars represent the standard deviation. Transcription initiated from 1H must be inferred by difference [1H – 1T] because the 1H primers also amplified 1T transcripts. Other abbreviations: Ce, corpus cerebelli; MO, medulla oblongata; OB, olfactory bulb; OT, optic tectum; SR, superior raphe; Te, telencephalon; TS, torus semicircularis. Scale bars: 250 µm (A,B,F-H); 1 mm (C,D). |
|
Gata1 binds distal and proximal promoter elements to regulate Hemogen expression in zebrafish. (A) Gata1 ChIP-sequencing showing enriched binding of Gata1 at CNE1 and CNE2 (C1 and C2, red lines) in the Hemogen promoter in adult zebrafish red blood cells (Yang et al., 2016). DNase-sequencing and ATAC-sequencing showing co-localization of the active chromatin regions (Yang et al., 2016). (B) Hemogen expression by WISH of wild-type (n=16/21) and (C) homozygous mutant (n=5/21) siblings (33 hpf) from in-crossed Gata1+/− vltm651 mutants. Insets show βe1-globin expression in mutant (n=4/10) and wild-type (n=6/10) siblings. Scale bar: 250 µm (C). |
|
Promoter elements have distinct roles in driving hematopoietic, renal and testicular expression of Hemogen in transgenic Tg(Hemgn:mCherry) zebrafish. (A) Schematic of the zebrafish Hemogen gene. CNEs, black; coding exons, white; transcription initiation sites, bent arrows. Three Tg(Hemgn:mCherry,myl7:EGFP) transgenes driven by portions of the Hemogen promoter were transfected into one-cell TU embryos by Tol2 transposase-mediated insertion. Numbers indicate length of promoter elements and arrows show gene direction. (B) 20 hpf. P1 transgene expression in the peripheral blood island (PBI). (C) 72 hpf. P1 transgene expression in the pronephric ducts (PD). (D) 5 dpf. P1 transgene expression in the proximal convoluted tubule (PCT). (E,F) 72 hpf. Co-localization of mCherry and EGFP in progenitors in the CHT of Tg(Hemgn-P1:mCherry,Lcr:GFP) or Tg(Hemgn-P1:mCherry,CD41:EGFP) zebrafish. (G) Transgene expression in mature erythrocytes from adult zebrafish. (H) Transgene expression in adult head kidney (HK), trunk kidney (TK) and tail kidney (T) near the EGFP+ heart (H). (I) Transgene expression in adult Sertoli cells (Se) that surround the seminiferous tubules (ST). (J) Proportion of embryos expressing transgenes P1, P2 or P3 in ICM, kidney, CHT and circulating primitive erythrocytes (RBC). Scale bars: 100 µm (B,D-F,I); 500 µm (C,H); 25 µm (G). EXPRESSION / LABELING:
|
|
Morpholino targeting of Hemogen inhibits erythropoiesis in embryonic zebrafish. Embryos were injected with 2 to 4 ng antisense MO targeted to the first 25 coding nucleotides of Hemogen. (A-B) O-dianisidine staining of erythrocytes was decreased in morphants (MO) relative to wild-type embryos (WT) or embryos rescued with 500 pg synthetic Hemgn mRNA (zHem) at 24 hpf. (ANOVA, Tukey post hoc test, P<0.001). (C-E) Live wild-type (C), Hem1 MO-injected (D) and Hem1 mm mismatch MO-injected (E) Tg(Lcr:EGFP)cz3325Tg embryos at 20 hpf. Morphants showed decreased EGFP expression in the ICM compared to the wild-type and mismatch MO controls. (F-H) Live wild-type (F), Hem1 MO-injected (G) and Hem1 mm MO-injected (H) embryos at 72 hpf. Morphant embryos have fewer EGFP+ cells in circulation compared to the two controls. The dorsal aortas of embryos (insets above F-H) were magnified 20× to permit quantitation of EGFP+ erythrocytes. Background red (D,G) and green (E,H) fluorescence was generated by the fluorescent labels on the MOs. (I) In vivo flow quantitation of EGFP+ erythrocyte concentrations between 3 and 6 dpf in Hem1-injected (n=9,7,7,7), Hem1mm-injected (n=13,14,11,11) and uninjected (n=5,10,10,9) embryos. Data shown as means±s.e.m. (*P≤0.05, **P≤0.001, ANOVA, Tukey-Kramer post hoc test). Arrowheads show notochord kinking. Scale bars: 500 µm (A-F); 100 µm (inset). EXPRESSION / LABELING:
PHENOTYPE:
|
|
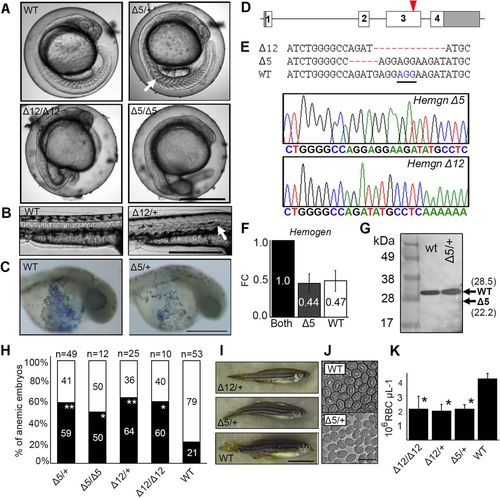
CRISPR/Cas9 mutagenesis of the third exon of zebrafish Hemogen reduces primitive and definitive erythropoiesis. Embryos were injected with Cas9 mRNA and a guide RNA to establish lines with mutations in exon three of zebrafish Hemogen. (A) 20 hpf. Representative wild-type and mutant siblings with notochord defects (arrow). (B) 48 hpf. Mutant Δ12 embryos with an in-frame deletion showing kinked notochords (arrow). (C) 24 hpf. Wild-type and Δ5/+ mutant embryos stained with diaminofluorene. Production of erythrocytes was reduced in heterozygotes. (D) Schematic of CRISPR/Cas9 target in the third exon (red arrowhead) of zebrafish Hemogen. (E) Sequences of founder mutations aligned at the CRISPR target site: Δ5 (Hemgnnuz2); Δ12 (Hemgnnuz4). The sequence traces show the Δ5 and Δ12 mutant alleles. PAM, blue and underlined; Δ, deletions (highlighted in red). (F) Relative expression of wild-type and Δ5 transcripts in blood from single adult, heterozygous Hemgnnuz2/+ mutants determined by qRT-PCR with allele specific primers. Three biological replicates were normalized to β-actin. Error bars represent the standard deviation. (G) Western blot of Hemogen in pooled 33 hpf wild-type embryos or pooled embryos from a Δ5 Hemgnnuz2/+ heterozygous in-cross. We calculated that the protein would run 6.5 kDa above its molecular weight at 28.5 kDa because of its high acidic composition (Guan et al., 2015). Arrows show the calculated sizes of wild-type and truncated alleles. (H) Proportion of genotyped mutants and wild-type sibling embryos at 2 dpf that were anemic (black) or phenotypically normal (white) (*P≤0.05, **P≤0.005, Chi square). (I) Wild-type and mutant zebrafish heterozygous for the Δ5 and Δ12 alleles. (J) Red blood cells from adult Hemgnnuz2/+ mutant zebrafish and wild-type siblings. (K) Erythrocyte counts in adult heterozygous Hemgnnuz2 (Δ5, n=12), heterozygous Hemgnnuz4 (Δ12, n=4) mutants, homozygous Hemgnnuz4 (Δ12, n=2) mutants and wild-type (n=9) siblings (*P≤0.05, ANOVA, Tukey post hoc test). Scale bars: 500 µm (A-C); 50 mm (I); 20 µm (J). PHENOTYPE:
|
|
Hemogen mutant zebrafish have increased cell death during embryonic development. (A) Genotypic ratios of 2 dpf embryos produced from heterozygous incrosses of Hemgnnuz3 (Δ5) or Hemgnnuz4 (Δ12) mutants. (B) Proportion of genotyped mutants and wild-type sibling embryos at 2 dpf that were apoptotic (black) or phenotypically normal (white) (* P ≤ 0.05, ** P ≤ 0.005, Chi square). (BC) Acridine orange staining for apoptotic cells is increased in the bodies and in the peripheral blood island (outlined) in 20 hpf heterozygous Hemgnnuz2 mutant zebrafish (n = 3) compared to wild-type siblings (n = 3). (D) Comparison of adult wild-type and homozygous Hemgnnuz4 (Δ12) mutant. (E) Average body length of adult wild-type and Hemgnnuz4 (Δ12) mutants. Error bars represent standard error (*, P ≤ 0.05, Student’s t test). Scale bars = 100 μm (E), 50 mm (D) PHENOTYPE:
|