- Title
-
Failed Progenitor Specification Underlies the Cardiopharyngeal Phenotypes in a Zebrafish Model of 22q11.2 Deletion Syndrome
- Authors
- Guner-Ataman, B., González-Rosa, J.M., Shah, H.N., Butty, V.L., Jeffrey, S., Abrial, M., Boyer, L.A., Burns, C.G., Burns, C.E.
- Source
- Full text @ Cell Rep.
|
Tbx1 Is Required for Pharyngeal Arch Artery, Head Muscle, and Outflow Tract Morphogenesis in Zebrafish (A–D) Confocal z-stacks of the head (A and C) and pharyngeal arch arteries (PAAs; B and D) in live 72 hr post-fertilization (hpf) control (CTRL; A and B; n = 60) and tbx1 mutant (C and D; n = 20) embryos carrying the Tg(kdrl:GFP) endothelial reporter. (B) and (D) are independent z-stacks acquired at higher magnification of the boxed regions in (A) and (C). Merged bright-field images and confocal z-stacks are shown in (A) and (C). The numbers in (B) identify each PAA. Lateral views, anterior left. (E–H) Bright-field z-stacks of the head (E and G) and pharyngeal arches (F and H) in 38 hpf CTRL (E and F; n = 40) and tbx1 mutant (G and H; n = 19) embryos processed by in situ hybridization with a tie1 riboprobe. The boxed regions in (E) and (G) are shown at higher magnification in (F) and (H). The numbers in (F) identify each tie1+ PAA angioblast cluster. Lateral views, anterior left. (I–L) Merged bright-field images and confocal z-stacks of the head region in 72 hpf CTRL (I and J; n = 40) and tbx1 mutant (K and L; n = 19) embryos co-immunostained with antibodies recognizing striated muscle (MF20, red) or OFT smooth muscle (Eln2, green). Ventral views, anterior up in (I) and (K). Lateral views, anterior left in (J) and (L). Arrow in (I) highlights the Eln2+ OFT smooth muscle that is missing in tbx1 mutants. In all cases, little to no variation was observed between animals in each experimental group. Scale bars, 25 μm. Pharyngeal arch1 (PA1)-derived head muscles: am, abductor mandibulae; do, dilator operculi; ima, intermandibularis anterior; imp, intermandibularis posterior; lap, levator arcus palatini; PA2-derived head muscles: ah, adductor hyoideus; ao, adductor operculi; hh, hyohyoidus; ih, interhyoidus; lo, levator operculi. LDA, lateral dorsal aorta; OFT, outflow tract; PAAs, pharyngeal arch arteries; V, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
tbx1 Mutant Embryos Lack nkx2.5+ Pharyngeal Progenitors (A and B) Merged bright-field and fluorescent images of 32 hr post fertilization (hpf) control (CTRL; A; n = 30) and tbx1 mutant (B; n = 8) embryos carrying the Tg(nkx2.5:ZsYellow) transgene. Dorsal views, anterior up. The numbers in (A) identify each pharyngeal cluster. (C and D) Bright-field z-stacks of 32 hpf CTRL (C; n = 51) and tbx1 mutant (D; n = 18) embryos processed by in situ hybridization with an nkx2.5 riboprobe. The numbers in (C) identify each pharyngeal cluster. Dorsal views, anterior up. (E and F) Merged bright-field and fluorescent images of 20 hpf CTRL (E; n = 30) and tbx1 mutant (F; n = 8) embryos carrying the Tg(nkx2.5:ZsYellow) transgene. The brackets in (E) highlight the ZsYellow+ the pharyngeal progenitor population missing in (F). Dorsal views, anterior up. (G and H) Bright-field z-stacks of 20 hpf CTRL (G; n = 32) and tbx1 mutant (H; n = 10) embryos processed by in situ hybridization with an nkx2.5 riboprobe. The brackets in (G) highlight the nkx2.5+ pharyngeal progenitor population missing in (H). Dorsal views, anterior up. In all cases, little to no variation was observed between animals in each experimental group. Scale bars, 25 μm. H, heart. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Tbx1 Is Required for Specification of the nkx2.5+ Pharyngeal Lineage in the ALPM of Zebrafish Embryos (A–H) Bright-field images of 2 somite stage (ss) (A, n = 30; E, n = 10), 6 ss (B, n = 60; F, n = 20), 10 ss (C, n = 30; G, n = 11), and 14 ss (D, n = 10; H, n = 10) control (CTRL) (A–D) and tbx1 mutant (E–H) embryos processed by in situ hybridization with an nkx2.5 riboprobe. Dorsal views, anterior up. (I and J) Bright-field z-stacks of 6 ss CTRL (I; n = 42) and tbx1 mutant (J; n = 13) embryos processed by in situ hybridization with a hand2 riboprobe. Dorsal views, anterior up. (K–N) Confocal z-stacks of CTRL (K and L) and tbx1 mutant (M and N) Tg(nkx2.5:Kaede) embryos imaged immediately following photoconversion at the 16 ss (K and M) and again at 32 hr post-fertilization (L and N). Four of four CTRL embryos contained photoconverted Kaede protein (KaedePC, pseudocolored yellow) in the heart and pharyngeal clusters at 32 hpf. Four of four mutant embryos contained KaedePC exclusively in the heart. Dorsal views, anterior up. In (A)–(K), little to no variation was observed between animals in each experimental group. Scale bars, 25 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
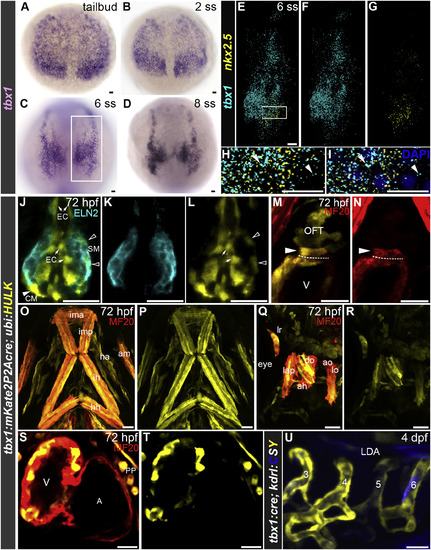
nkx2.5+ Progenitors in the ALPM of Zebrafish Embryos Co-express tbx1 (A–D) Bright-field images of wild-type embryos (n = 15 per stage) between tail bud and 8 somite stage (ss) processed by in situ hybridization with a tbx1 riboprobe. Boxed area in (C) shows region of embryo analyzed in (E)–(G). Dorsal views, anterior up. (E–I) Confocal z-stack of the tbx1 and nkx2.5 expression domains on the right side of a 6 ss wild-type embryo (n = 14) processed by RNAscope double-fluorescent in situ hybridization with tbx1 (pseudocolored cyan) and nkx2.5 (pseudocolored yellow) riboprobes. Merged double- (E) and single-channel (F and G) images are shown. Projection of selected z-stack images from the boxed region in (E) is shown at higher magnification in (H). A single section from (H) is shown in (I) (arrowheads are shown to indicate regions of overlap) with DAPI nuclei counterstaining. Dorsal views, anterior up. (J–T) Single optical sections (J–N, S, T) or confocal z-stacks (O–R) of the OFT (J–N), PA1- and PA2-derived HMs (O–R), and heart (S and T) in a 72 hpf Tg(tbx1:mKate2P2Acre), Tg(ubi:HULK) double-transgenic embryo co-immunostained with antibodies recognizing GFP (pseudocolored yellow) and OFT smooth muscle (Eln2; J–L; pseudocolored cyan, n = 15) or GFP (pseudocolored yellow) and striated muscle (MF20; M–T; red, n = 39) antibodies. Arrows, open arrowheads, and closed arrowheads point to GFP+ OFT endothelial cells (EC), OFT smooth muscle cells (SM), and OFT cardiomyocytes (CM) in (J), (L), and (M), respectively. Dashed lines demarcate the boundary between the ventricular and OFT myocardium in (M) and (N). Ventral views, anterior up (J–P, S, T). Lateral view, anterior left (Q and R). Merged (J, M, O, Q, S) and single (K, L, N, P, R, T)-channel images are shown. (U) Confocal z-stack of a 4 days post-fertilization (dpf) Tg(tbx1:mKate2P2Acre); Tg(kdrl:CSY) double-transgenic embryo (n = 22) showing expression of AmCyan and ZsYellow from the unrecombined and recombined reporter, respectively. Lateral view, anterior to the left. The numbers identify each PAA. In (J)–(N and (S)–(U), all of the animals contained some reporter expression in the structures shown, but the distribution within each structure was variable. In (O) and (P), little to no variation was observed in head muscle labeling between animals. In (A)–(I), little to no variation was observed between animals in each experimental group. Scale bars, 25 μm. PA1-derived head muscles: am, abductor mandibulae; do, dilator operculi; ima, intermandibularis anterior; imp, intermandibularis posterior; lap, levator arcus palatini; PA2-derived head muscles: ah, adductor hyoideus; ao, adductor operculi; ha, hypobranchial artery; hh, hyohyoidus; ih, interhyoidus; lo, levator operculi; lr, lateral rectus ocular muscle. A, atrium; LDA, lateral dorsal aorta; OFT, outflow tract; PP, parietal pericardium; V, ventricle. EXPRESSION / LABELING:
|
|
gdf3 Is Downstream of tbx1 during Specification of nkx2.5+ Pharyngeal Progenitors (A) Bar graph showing the relative expression levels of gdf3, nkx2.5, and tbx1 transcripts in 6 somite stage (ss) control (CTRL) and tbx1 mutant embryos measured by qPCR. Three biological replicates were analyzed. Error bars show SD. ∗p < 0.05 as determined by an unpaired two-tailed t test. (B and C) Bright-field images of 6 ss CTRL (B; n = 40) and tbx1 mutant (C; n = 12) embryos processed by in situ hybridization with a gdf3 riboprobe. Dorsal views, anterior up. (D–I) Confocal z-stacks of the tbx1, gdf3, and nkx2.5 expression domains on the right side of a 6 ss wild-type zebrafish embryo (n = 7) processed by RNAscope triple-fluorescent in situ hybridization with tbx1 (pseudocolored cyan), gdf3 (pseudocolored magenta), and nkx2.5 (pseudocolored yellow) riboprobes. Merged triple- (D), double- (E and F), and single- (G–I) channel images are shown. (J) Projection of selected z-stack images from the boxed region in (D) is shown at higher magnification. (K) A single confocal section from (J) is shown in (K) (arrowheads are shown to indicate regions of overlap) with DAPI nuclei counterstaining. (L) Schematic diagrams showing the domain structures of the predicted protein products of wild-type (WT) gdf3 and a null allele of gdf3, gdf3fb20, generated by CRISPR/Cas9-mediated genome editing. The asterisk highlights the location of a frameshifting 10-bp deletion in the gdf3fb20 mRNA, which causes the translation of six ectopic amino acids (white) before a premature stop codon truncates the protein upstream of the mature ligand domain. (M–O) Bright-field z-stacks of live 28 hr post-fertilization (hpf) CTRL (M; n = 25), zygotic gdf3 (N; Zgdf3, n = 25), and maternal-zygotic gdf3 (O; MZgdf3, n = 40) zebrafish embryos. Lateral views, anterior left. In all cases, little to no variation was observed between animals in each experimental group. Scale bars, 25 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Small-Molecule-Mediated Inhibition of the Gdf3 Receptor Alk4 Phenocopies the Pharyngeal Defects Observed in tbx1 Mutants (A) Experimental timeline of SB-505124 treatment and phenotypic analyses. (B–D) Bright-field z-stacks of 38 hr post-fertilization (hpf) wild-type embryos treated with DMSO or SB-505124 between 75% epiboly and 24 hpf, and processed by in situ hybridization with a tie1 riboprobe. Images of representative animals with unaffected (B; 3 or more clusters), reduced (C; 1–2 clusters), or absent (D; 0 clusters) tie1+ pharyngeal arch artery (PAA) clusters are shown. The numbers in (B) and (C) identify each PAA cluster. The asterisks highlight clusters of smaller size in (C). Lateral views, anterior left. (E) Bar graph showing the percentages of embryos with unaffected, reduced, or absent PAA clusters after treatment with DMSO (n = 22), 40 μM SB-505124 (n = 27), or 80 μM SB-505124 (n = 12). (F–K) Confocal z-stacks of 72 hpf wild-type embryos treated between 75% epiboly and 24 hpf with DMSO or SB-505124 and co-immunostained with antibodies recognizing striated muscle (MF20, red) or outflow flow tract (OFT) smooth muscle (Eln2, green). Images of representative animals with unaffected (F and I), reduced (G and J), or absent (H and K) head muscles and OFTs (white arrows, F and G) are shown. (F–H) Ventral views, anterior up. (I–K) Lateral views, anteior left. (L and M) Bar graphs showing the percentages of animals with unaffected, reduced, or absent head muscles (L) or OFTs (M) after treatment with DMSO (n = 49), 20 μM SB-505124 (n = 40), or 40 μM SB-505124 (n = 53). Scale bars, 25 μm. PA1-derived head muscles: am, abductor mandibulae; do, dilator operculi; ima, intermandibularis anterior; imp, intermandibularis posterior; lap, levator arcus palatini; PA2-derived head muscles: ah, adductor hyoideus; ao, adductor operculi; hh, hyohyoidus; ih, interhyoidus; lo, levator operculi. LDA, lateral dorsal aorta. OFT, outflow tract; V, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Small-Molecule-Mediated Inhibition of the Gdf3 Receptor Alk4 Phenocopies the nkx2.5+ Progenitor Specification Defect Observed in tbx1 Mutants (A–C) Bright-field images of 6 somite stage (ss) wild-type embryos processed by in situ hybridization with an nkx2.5 riboprobe after being treated from 75% epiboly with DMSO (A; n = 18), 20 μM SB-505124 (B; n = 17), or 40 μM SB-505124 (C; n = 20). Dorsal views, anterior up. (D) Dot plot showing the average sizes of the bilateral nkx2.5 expression domain, normalized to the total embryo area shown in each image, in each experimental group. ∗∗p < 0.01; ∗∗∗∗p < 0.0001, as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. (E–G) Bright-field images of 6 ss wild-type embryos processed by in situ hybridization with a tbx1 riboprobe after being treated with DMSO (E; n = 18), 20 μM SB-505124 (F; n = 18), or 40 μM SB-505124 (G; n = 19) beginning at 75% epiboly. Dorsal views, anterior up. (H) Dot plot showing the average sizes of the bilateral tbx1 expression domain, normalized to the total embryo area shown in each image, in each experimental group. n.s., nonsignificant as determined by a one-way ANOVA test. Scale bars, 25 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
tbx1 mutants do not display elevated rates of apoptosis in anterior lateral plate mesoderm, Related to Figure 3. (A, B) Confocal z-stacks of 6 somite stage (ss) control (CTRL; A, n=14) and tbx1 mutant (B, n=8) Tg(tbx1:GFP) embryos processed for TUNEL labeling (red), immunostained to detect GFP (green), and counterstained with DAPI (blue). The boxed regions encompass the approximate locations of nkx2.5+ progenitors. Dorsal views, anterior up. (C, D) Box and whisker plots showing the distribution of total apoptotic cells (C) or apoptotic cells within the boxed areas (D) per embryo in each experimental group. n.s., non significant as determined by unpaired, two-tailed t-test. Scale bars=25mm. EXPRESSION / LABELING:
|
|
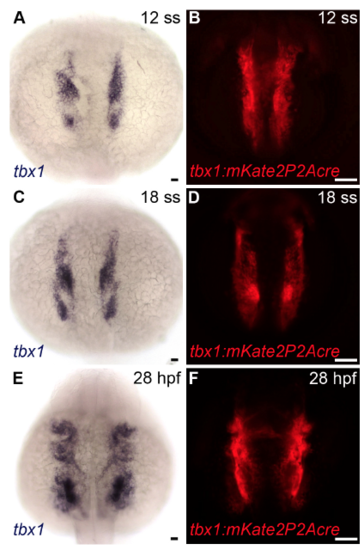
Recapitulation of the tbx1 expression pattern by mKate2 in Tg(tbx1:mKate2P2Acre) animals, Related to Figure 4. (A-F) Brightfield (A, n=15; C, n=14; E, n=15) and fluorescent (B,D,F, n=40) images of 12 somite stage (ss). (A,B), 18 ss (C,D) and 28 hours post-fertilization (hpf) (E,F) wild-type zebrafish embryos processed by in situ hybridization with a tbx1 riboprobe (A,C,E) or live Tg(tbx1:mKate2P2Acre) animals imaged in the red channel. Dorsal views, anterior up. Little to no variation was observed between animals in each experimental group. Scale bars=25μm. EXPRESSION / LABELING:
|
|
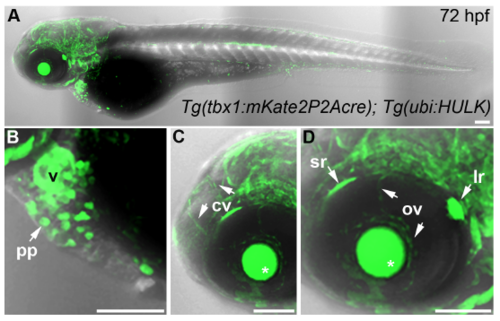
Genetic lineage tracing of tbx1+ cells, Related to Figure 4. (A-D) Merged brightfield images and confocal z-stacks of 72 hours post-fertilization (hpf) Tg(tbx1:mKateP2Acre), Tg(ubi:HULK) double transgenic embryos (n=40) immunostained to detect GFP. Higher magnification views of (A) are shown in (B), (C), and (D). White arrows highlight labeled cells in the parietal pericardium (pp), cranial vasculature (cv), superior rectus (sr) ocular muscle, lateral rectus (lr) ocular muscle, and ocular vasculature (ov). Fluorescence in the lens (*) results from a cry:GFP selectable marker on the Tg(ubi:HULK) vector backbone. Lateral views, anterior left. Little to no variation was observed between animals in each experimental group. Abbr: v, ventricle. Scale bars=100 μm. EXPRESSION / LABELING:
|
|
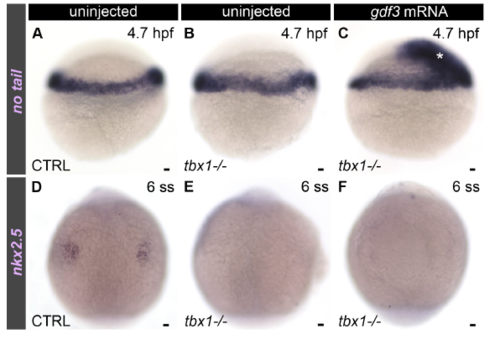
Injection of gdf3 mRNA into tbx1 mutants is not sufficient to rescue nkx2.5 expression at 6 somite stage, Related to Figure 5. (A-F) Brightfield images of 4.7 hours post-fertilization (hpf) (A-C) or 6 somite stage (ss) (D-F) uninjected control (CTRL; A, n=28; D, n=29) and tbx1 mutant (B, n=7; E, n=8) embryos or tbx1 mutant embryos injected with gdf3 mRNA at the one-cell stage (C, n=6; F, n=8). Embryos were processed by in situ hybridization with no tail (A-C) or nkx2.5 riboprobes (D-F). The asterisk in (C) highlights ectopic mesoderm induced by gdf3 mRNA. Lateral views in (A-C). Dorsal views, anterior up in (D-F). Little to no variation was observed between animals in each experimental group. Scale bars=25μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
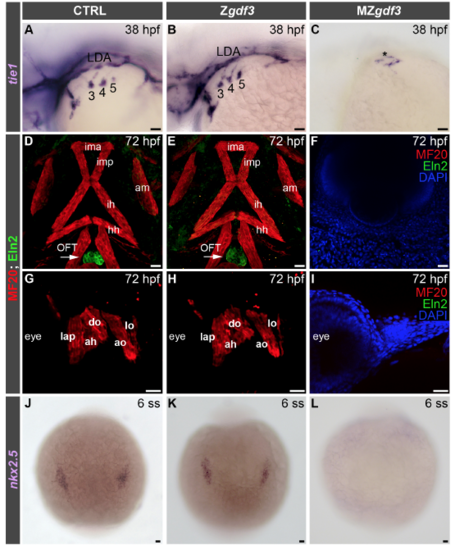
Analysis of zygotic and maternal zygotic null gdf3 embryos, Related to Figure 5. (A-C) Brightfield images of 38 hours post-fertilization (hpf) control (CTRL; n=20), zygotic null gdf3 (Zgdf3; n=7), and maternal zygotic null gdf3 (MZgdf3; n=25) embryos processed by in situ hybridization with a tie1 riboprobe. The numbers identify each PAA cluster in (A) and (B). The asterisk in (C) highlights endothelial cells of unknown identity. Lateral views, anterior left. (D-I) Confocal z-stacks of 72 hpf CTRL (n=14), Zgdf3 (n=11), and MZgdf3 (n=32) embryos double immunostained with antibodies recognizing striated muscle (MF20, red) and OFT smooth muscle (Eln2, green). Arrows in (D,E) highlight the Eln2+ OFT smooth muscle that is missing in MZgdf3 mutants. Nuclei were counterstained with DAPI in (F,I). Merged double (D,E,G,H) and triple channel (F,I) images are shown. MZgdf3 animals are devoid of head and trunk mesoderm and therefore lack striated and smooth muscle. Ventral views, anterior up in (D-F). Lateral views, anterior left in (G-I). (J-L) Brightfield images of 6 somite stage (ss) CTRL (n=21), Zgdf3 (n=6), and MZgdf3 (n=26) embryos processed by in situ hybridization with an nkx2.5 riboprobe. Dorsal views, anterior up. In all cases, little to no variation was observed between animals in each experimental group. Abbr: LDA, lateral dorsal aorta; PA1-derived head muscles: ima, intermandibularis anterior; imp, intermandibularis posterior; am, abductor mandibulae; lap, levator arcus palatini; do, dilator operculi; PA2- derived head muscles: ih, interhyoidus; hh, hyohyoidus; ah, adductor hyoideus; ao, adductor operculi; lo, levator operculi; OFT, outflow tract. Scale bars=25 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
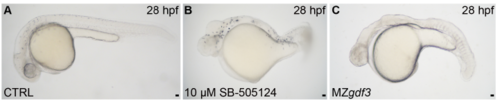
Antagonizing the Gdf3 receptor Alk4 with small molecule SB-505124 phenocopies MZgdf3 animals, Related to Figure 6. (A,B) Brightfield images of 28 hours post-fertilization (hpf) wild-type animals treated with DMSO (CTRL, A, n=25) or 10mM SB-505124 (B, n=25) and a 28 hpf MZgdf3 embryo (C, n=40). Lateral views, anterior left. Little to no variation was observed between animals in each experimental group. Scale bars=25μm. PHENOTYPE:
|