- Title
-
In vivo analysis of cardiomyocyte proliferation during trabeculation.
- Authors
- Uribe, V., Ramadass, R., Dogra, D., Rasouli, S.J., Gunawan, F., Nakajima, H., Chiba, A., Reischauer, S., Mochizuki, N., Stainier, D.Y.R.
- Source
- Full text @ Development
|
Tg(myl7:mVenus-Gmnn) expression marks proliferating cardiomyocytes in zebrafish. (A-F) Distribution of myl7:mVenus-Gmnn+ cardiomyocytes at several developmental stages. Tg(myl7:MKATE-CAAX) expression labels cardiomyocyte membranes. (A-C,E) Three-dimensional reconstructions; (D,F) cross-sections through the ventricle; white arrows point to trabecular mVenus-gmnn+ cardiomyocytes. (G) Cross-section of a 102 hpf heart stained for EdU and myosin heavy chain (MHC), to mark cardiomyocytes, after 6 h of EdU incubation. The location of EdU+ cardiomyocytes in the trabeculae is similar to that of myl7:mVenus-Gmnn+ cardiomyocytes (F,G; red arrowheads). (H) Cross-section of a 120 hpf Tg(myl7:nls-DsRed) heart stained for EdU, MHC and DsRed after 24 h of EdU incubation. Red arrowheads point to EdU+ cardiomyocytes in the trabecular layer. (I) Three-dimensional reconstruction of a 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:nlsDsRed) heart. (J) Three-dimensional reconstruction of a 102 hpf Tg(myl7:nlsDsRed) heart stained for EdU and DsRed after 6 h of EdU incubation. (K) Percentage of proliferating cardiomyocytes at several developmental stages, as assessed with the Tg(myl7:mVenus-gmnn) line or EdU labeling (6 h) (data are presented as mean±s.d.). (L) Three-dimensional reconstruction of a 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:mCherry-cdt1) heart that was temporarily stopped to follow the progression of the cardiomyocyte cell cycle. (M) Higher magnification of the ventricular area of the 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:mCherry-cdt1) heart shown in L followed over 10 h. White arrowheads point to a cardiomyocyte that transits from a myl7:mVenus-Gmnn−; myl7:mCherry-Cdt1+ state at t=0:00 to a myl7:mVenus-Gmnn+; myl7:mCherry-Cdt1− state, with completed nuclear division at t=8:00 h. White arrows point to a cardiomyocyte that has started expressing myl7:mVenus-Gmnn at t=0:00 h and completes nuclear division at t=7:00 h. Yellow arrowheads point to additional examples of dividing cardiomyocytes (from t=5:00 to t=7:00); yellow arrows point to a cardiomyocyte losing mVenus-Gmnn signal. (N) Estimation of the duration of the S-G2-M phase (green) based on time-lapse imaging. Scale bars: 20 µm (bar in J applies to I). EXPRESSION / LABELING:
|
|
Behavior of myl7:mVenus-Gmnn+ cardiomyocytes at early stages of trabecular formation. (A-C) Distribution of myl7:mVenus-Gmnn+ cardiomyocytes in the compact and trabecular layers at 72, 82 and 96 hpf. Tg(myl7:MKATE-CAAX) expression labels cardiomyocyte membranes; white arrows point to trabecular cardiomyocytes. (D) Percentage of myl7:mVenus-Gmnn+ cardiomyocytes in the compact and trabecular layers relative to the total number of myl7:mVenus-Gmnn+ cardiomyocytes at the corresponding developmental stages. (E) Number of myl7:mVenus-Gmnn+ cardiomyocytes in the compact and trabecular layers at 72 and 82 hpf; eight larvae analyzed at 72 hpf, nine larvae at 82 and 96 hpf. Results are mean±s.d., **P=0.0089 and 0.0021; Student's two tailed t-test. (F-H) Time-lapse imaging of Tg(myl7:mVenus-gmnn); Tg(myl7:mCherry-CAAX) beating hearts from 72 (F,G) or 82 (H,I) hpf. (F) Parallel divisions are observed in the compact layer (white arrowheads) (six larvae imaged, five compact layer divisions observed). (G) Delamination of myl7:mVenus-Gmnn+ cardiomyocyte (six larvae imaged; four myl7:mVenus-Gmnn+ cardiomyocytes were observed to be delaminating). White arrowheads point to delaminating mVenus-gmnn+ cardiomyocytes. (H) Divisions in the trabecular layer (yellow arrowheads) can occur in an orientation parallel to the compact layer (five larvae imaged; eight parallel divisions observed in the trabecular layer and seven in the compact layer). (I) Time-lapse live imaging of temporarily stopped Tg(myl7:mVenus-gmnn); Tg(myl7:MKATE-CAAX) hearts from 82 hpf using lightsheet microscopy. The myl7:mVenus-Gmnn signal becomes brighter before division (red arrowheads; red dashed line outlines dividing cardiomyocyte), and when the nuclear membrane breaks down during the early stages of mitosis, the fluorescent signal can be detected throughout the cell (t=0:30); however, it relocalizes to the nucleus when cytokinesis is completed (t=1:30); three larvae imaged. Images on the right are higher magnifications of the boxed area on the image on the left. Time is in h:min. Scale bars: 20 µm. EXPRESSION / LABELING:
|
|
Sarcomere disassembly, as well as cell shape and volume changes, are observed before cardiomyocyte division. (A-C) Three-dimensional reconstruction of 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:BFP-CAAX) larvae injected with myl7:mScarlet plasmid to label individual cardiomyocytes; (D-F′) the cardiomyocytes used for 3D surface rendering are outlined in A-C. (D-F′) Three-dimensional surface rendering of individual mitotic (D-E′) and interphase (F,F′) cardiomyocytes. The thickness of the mitotic cardiomyocytes is increased and their shape appears more rounded. (G) Measurement of the cell volume of mitotic and interphase cardiomyocytes located in the outer curvature of the heart, as measured through the mVenus-Gmnn or mScarlet signal; each point represents a cardiomyocyte from seven different larvae. Data are mean±s.d., *P=0.0266; Student's two-tailed t-test. (H) Time-lapse imaging of beating Tg(myl7:actn3b-EGFP); Tg(myl7:mCherry-CAAX) hearts starting at 80 hpf. Dashed line outlines the dividing cardiomyocyte. The organized sarcomeric pattern (a,a′) becomes gradually lost (b-c′) and the Actn3b-EGFP signal is detected close to the borders of the cardiomyocyte (d,d′) prior to its division (e,e′). Five larvae imaged; four cardiomyocytes were seen completing their division. (I-J) Three-dimensional reconstruction of 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:LA-tdTomato); Tg(myl7:BFP-CAAX) hearts (12 larvae imaged). I′ is a mirror image of I. (K) Higher magnification of the ventricular region outlined in I′. An individual interphase cardiomyocyte exhibits an organized striated pattern of sarcomeres (K′), whereas in a dividing cardiomyocyte, the organized pattern is lost and the fluorescent signal appears more diffuse (K″). (L,L′) Higher magnification of the atrial area outlined in J, where sarcomere disassembly is clearly visible in a dividing cardiomyocyte (L′). (M-M″) Details of a dividing cardiomyocyte in the atrium that appears to be undergoing cytokinesis. Organized sarcomeric structures are absent, in contrast to neighboring cardiomyocytes (M″). Scale bars: 20 µm in A,H,I,K,L,M; 5 µm in D,K′. Bars in I,K′,L and M apply to I-J,K″,L′ and M′,M″. EXPRESSION / LABELING:
|
|
Erbb2 signaling controls cardiomyocyte proliferation and trabecular growth. (A-C′) Treatment of Tg(myl7:mVenus-gmnn); Tg(myl7:MKATE-CAAX) larvae with DMSO control or the Erbb2 inhibitor PD168393 from 96 to 108 hpf (A and B show 3D reconstruction of control and treated hearts; A′ and B′ show cross-sections). (C) Quantification of the total number of myl7:mVenus-Gmnn+ cardiomyocytes and (C′) the number of myl7:mVenus-Gmnn+ cardiomyocytes in the compact and trabecular layers after 12 h of treatment. There is a decrease in the total number of proliferative cardiomyocytes, particularly in the trabecular layer; each point represents a heart. (D-F″) Images of 10 dpf Tg(myl7: H2B-EGFP); Tg(myl7:MKATE-CAAX) larvae that have been treated with DMSO control or PD168393 from 96 to 108 hpf. The number of cardiomyocytes observed in the trabecular layer is reduced (compare D′ with E′), and a simplified trabecular network is formed in PD168393-treated larval hearts (compare 3D surface rendering D″ with E″). In the most severe cases, trabeculae do not project into the lumen, and trabecular cardiomyocytes stay close to the compact layer (compare D′,D″ with F′,F″). (G,H) Quantification of the number of cardiomyocytes in control and PD168393-treated larvae. While the number of compact layer cardiomyocytes remains unchanged, the number of trabecular cardiomyocytes is significantly reduced (G), resulting in an overall reduction in the number of cardiomyocytes in treated hearts. Each point represents a heart. (I) The body length of these larvae is not affected (each point represent a larva). (J,K) Phospho-histone 3 (PH3) staining of 48 hpf Tg(myl7:nrg2a-p2a-tdTomato) hearts. More PH3+ cardiomyocytes are observed in nrg2a-overexpressing hearts compared with control and some cardiomyocytes divide from the compact layer towards the lumen (K, inset). (L-S) Hearts incubated with EdU for 24 h, from 48 to 72 hpf (L-O) or from 72 to 96 hpf (P-S), and stained for PH3. Divisions perpendicular to the compact layer towards the lumen are detected (M, details) in nrg2a-overexpressing cardiomyocytes, in contrast to control, where only parallel divisions can be observed (L, inset). An increased rate of proliferation in nrg2a-overexpressing cardiomyocytes is observed in both compact and non-compact layers; each point represents a heart (N,O,R,S). Data are mean±s.d., ****P<0.0001 in C,C',G,N,O,R,S, *P=0.0122 in C′, **P=0.0033 in H, **P=0.0056 in O, ***P=0.0003 in S; Student's two-tailed t-test. Experiments in A-C',J-S were repeated twice, experiments in D-I were repeated three times. Scale bars: 20 µm. Scale bars in J,L,P apply to K,M,Q, respectively. |
|
The TGFβ pathway ligand Inhbaa promotes trabecular cardiomyocyte proliferation. (A-H) Quantification and images of cardiomyocyte proliferation in the Tg(myl7:inhbaa-2A-H2B-EGFP) line after EdU incubation from 72 to 96 hpf (A-D) and from 96 to 120 hpf (E-H) in the two myocardial layers; myosin heavy chain staining labels cardiomyocytes. Overexpression of inhbaa drives cardiomyocyte proliferation in the trabecular, but not the compact, layer; each point represents a heart (D,H). (I-J″) Evaluation of the trabecular phenotype in the Tg(myl7:inhbaa-2A-H2B-EGFP) line at 10 dpf; Tg(myl7:MKATE-CAAX) expression labels cardiomyocyte membranes. A denser trabecular network is observed in inhbaa-overexpressing cardiomyocytes (compare cross-section in I′ with J′, and 3D surface rendering I″ with J″). Six control and seven inhbaa-overexpressing larvae are imaged. Data are mean±s.d., *P=0.041 in C, **P=0.0025 in D, **P=0.0011 in G, ***P=0.0007 in H; Student's two-tailed t-test; each experiment was repeated twice. Scale bars: 20 µm. Scale bars in A,E,I,J apply to B,F,I′,J′, respectively. |
|
myl7:mVenus-Gmnn signal diffuses throughout the cardiomyocyte prior to cytokinesis. Zoomed image of an atrial cardiomyocyte in a 82 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:MKATE-CAAX) heart acquired using a lightsheet microscope time-lapse for every 30 minutes. Diffusion of the fluorescent signal throughout the cardiomyocyte is observed before it divides. Once cytokinesis is completed, the myl7:mVensu-Gmnn signal is again restricted to the nuclei and then disappears within 1 hour. Scale bars, 20 µm. |
|
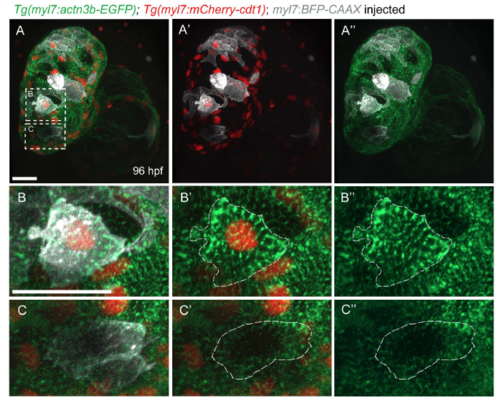
Sarcomere organization in G1 cardiomyocytes. (A-A") Three-dimentional reconstruction of a 96 hpf Tg(myl7:actn3b-EGFP); Tg(myl7:mcherry-cdt1) heart injected with myl7:BFP-CAAX plasmid to label individual cardiomyocytes. (B-B") Close-up of a cardiomyocyte in G1 phase, as labeled by mCherry+ nucleus; note the continuous sarcomeric striations throughout the cardiomyocyte. (C-C") Close-up of a myl7:mCherry-cdt1- cardiomyocyte; the sarcomeres do not occupy the whole cardiomyocyte, but are mostly located at the periphery of the cell. Scale bars, 20 µm. |
|
Sarcomere disassembly during cardiomyocyte division. Time-lapse live imaging starting from 80 hpf of a Tg(myl7:actn3b-EGFP); Tg(myl7:nlsDsRed) heart. (A-A") At t=0:00, striated sarcomeres were observed in all cardiomyocytes. (B-B") A cardiomyocyte is dividing (white arrow), and the EGFP signal is seen outlining the borders of the two nascent daughter cardiomyocytes. (C-C") Two daughter cells can be observed, but still the sarcomeres remain on the periphery of these new cardiomyocytes. (D-F") A recovery of the striated pattern in the newly formed cardiomyocytes is observed. (G) Schematic representation of the key time points of the sarcomere disassembly during cardiomyocyte division. Scale bars, 20 µm. |
|
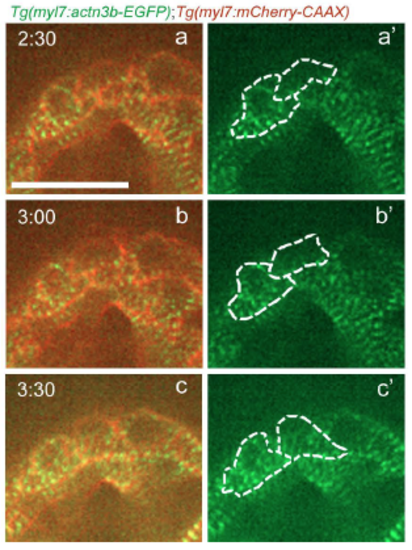
Sarcomere reassembly after cardiomyocyte cytokinesis. Follow up of the time-lapse live imaging of the beating Tg(myl7:actn3b-EGFP); Tg(myl7:mCherry-CAAX) heart shown in Figure 3H. The two daughter cells reassemble their sarcomeres (a-b'), and seem to recover the striated sarcomeric pattern approximately 3 hours after the disassembly was first observed in the dividing cardiomyocyte (c-c'). Scale bar, 20 µm. |
|
inhibition of Erbb2 signaling at different larval stages leads to reduced cardiomyocyte proliferation. (A-C') 96 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:MKATE-CAAX) larvae after treatment with PD168393 from 72 to 84 hpf. Upon treatment, the overall number of proliferative cardiomyocytes in the ventricle is decreased (C), affecting both compact and trabecular layers (C'). Each point represents a heart. (D-F") 144 hpf Tg(myl7:mVenus-gmnn); Tg(myl7:MKATE-CAAX) larvae after treatment with PD 168393 from 96 to 108 hpf. The number of dividing cardiomyoctyes is reduced in the treated larvae (F), and the effect on the trabecular layer is more pronounced (F'). Each point represents a heart. (J-L') 144 hpf Tg(myl7:nlsDsRed) larvae after treatment with PD168393 from 96 to 108 hpf, and incubated with EdU in parallel. Immunostaining against heavy chain myosin and DsRed was performed to label membranes and cardiomyocyte nuclei, respectively. The percentage of EdU* cardiomyocytes is reduced (L). du to a lower proliferation rate in both myocardial layers (L'); each point represents a heart. Results are mean S.D., ****P<0.0001; **P=0.0018 in F', ***P=0.0005 in I, **P=0.0056 and **P=0.0034 in I', ***P=0.0005 in L, *P=0.034 and ***P=0.0002in L"; Students two-tailed t-test; experiment was repeated two times. Scale bars, 20 µm |
|
Effects of Erbb2 signaling inhibition at later larval stages. (A-D) 10 dpf Tg(myl7:H2B-EGFP); Tg(myl7:MKATE-CAAX) larvae after treatment with PD168393 from 72 to 84 hpf. Overall, little difference is observed between control and treated hearts, as can be seen from both optical sections (compare A' to B') and 3d surface renderings (compare A" to B"). The numbers of compact layer cardiomyocytes are similar, while the numbers of trabecular layer cardiomyocytes are slightly, although not significantly, increased (C) and the total numbers of cardiomyocytes are similar in both groups (D); each point represents a heart. (E-H) 1- dpf Tg(myl7:H2B-EGFP); Tg(myl7:MKATE-CAAX) larvae after treatment with PD168393 from 120 to 132 hpf. The trabeculae appear shorter and the trabecular mesh looks less developed than in control (compare E',E" to F',F"). The number of trabecular cardiomyocytes is slightly reduced in the treated larvae (G), but the total number of cardiomyocytes is similar (H); each point represents a heart. (I-J) Body length of 10 dpf larvae after treatment with PD168393 from 72 to 84 hpf (I) and from 120 to 132 hpf (J); each point represents a larva. Results are mean S.D., P=0.073 in G trabecular layer, *P=0.044 in J, Student's two-tailed t-test; experiment was repeated three times. Scale bars, 20 µm. |
|
Increased trabeculation in Tg(myl7:inhbaa-2AH2B-EGFP) hearts at early larval stages. (A-C') 96 hpf Tg(myl7:MKATE-CAAX) control larva (A-A') and Tg(myl7:inbaa-2A-H2B-EGFP); Tg(myl7:MKATE-CAAX) larva with no obvious phenotype (B-B') or with signs of hypertrabeculation (C-C'). (D-E') 120 hpf Tg(myl7:MKATE-CAAX) control larva (D-D') and Tg(myl7:inhbaa-2A-H2B-EGFP), Tg(myl7:MKATE-CAAX) larva where increased trabeculation can be observed E-E'). Scale bars, 20 µm. |
|
mstnb mutants exhibit increased cardiomyocyte proliferation in both compact and trabecular layers. (A) images of 120 hpf Tg(myl7:nlsDsRed) mstnb+/- and mstnb-/- larcae stained for EdU, MHC and DsRed after 24 hours of EdU incubation. In the mutants, both compact and trabecular cardiomyocytes exhibit a moder increase in their proliferative rates (results are mean S.D., compact layer, *P=0.036, trabecular layer, *P=0.045; total number, *P=0.0301; Student's two-tailed t-test; experiment was repeated 3 times). (B) pSmad3 immunostaining on 120 hpf wild-type hearts; myosin heavy chain staining labels cardiomyocytes. (C-C') Close-up of the outer curvature of the ventricle, where trabecular (yellow arrowheads) and compact (purple arrowhead) pSmad+ cardiomyocytes are observed. (D) Quantification of the percent of pSmad+ cardiomyocytes in the compact and trabecular layers; each point represents a heart. *P=0.01; Student's two-tailed t-test; experiment was repeated two times. Scale bars, 20 µm. |