- Title
-
A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish
- Authors
- Wu, R.S., Lam, I.I., Clay, H., Duong, D.N., Deo, R.C., Coughlin, S.R.
- Source
- Full text @ Dev. Cell
|
G0 Mutation of s1pr2 by Single- or Four-Guide Cas9 RNP (A) Embryos were cell-injected at the one-cell stage with single-guide or four-guide Cas9 RNP targeting s1pr2 and scored for phenotypes at 1 day post fertilization (dpf) and 4 dpf. Embryos were first scored as dysmorphic or not (lower panel). Non-dysmorphic embryos were then scored for the miles apart cardia bifida phenotype as defined in Figure S1. The percentage with strong (blue), intermediate (red), or no (green; wild-type [WT]) phenotype is indicated. The number of non-dysmorphic embryos scored is shown at bottom. ∗∗∗∗p < 0.0001; ns, not significant; Fisher's exact test for Strong versus other phenotype. None of 76 embryos injected with control sgRNA exhibited cardia bifida; 5% were dysmorphic. The figure represents pooled data from two independent experiments. (B) Representative Tg(myl7:GFP) G0 embryo with a Strong cardia bifida phenotype (bottom) after injection of a four-guide Cas9 RNP mix. A control clutchmate is shown at the top. Tg(myl7:GFP) labels the heart fields (green). Scale bar, 250 μm. (C and D) Embryos injected with a four-guide Cas9 RNP mix targeting s1pr2 were collected at 1 dpf. DNA was prepared from two pools of 3 embryos (two separate experiments); s1pr2 amplicons were cloned and 30 were sequenced. (C) Wild-type (top line) and ten representative mutant sequences from injected embryos. Arrows denote predicted site of double-stranded breaks corresponding to each of the four guides used in (A). Sequences predicted to hybridize with CRISPR sgRNA spacers are highlighted with alternating green and yellow shading. (D) Summary of characteristics of 30 s1pr2 alleles analyzed. |
|
Onset and Durability of Phenotypes in G0 Embryos and Adults Embryos were cell-injected at the one-cell stage with four-guide Cas9 RNP targeting casanova (sox32) (A–D) or tyr (E) or with scrambled guides as indicated. (A–D) Impaired endoderm marker expression in G0 embryos after four-guide targeting of sox32. (A and B) Fish hemizygous for the endoderm marker Tg(sox17:GFP) were intercrossed and the resulting embryos injected with the indicated four-guide Cas9 RNP and imaged at 10 hpf. (A) Representative images. Scale bar, 250 μm. (B) Percentage of embryos expressing GFP (green) or not (blue). Dotted line indicates 75%, the percentage of fluorescent embryos expected from the hemizygous transgenic cross. ∗∗∗∗p < 0.0001; ns, not significant; Fisher's exact test for presence or absence of GFP fluorescence. The number of embryos analyzed is shown in each column. (C) Embryos injected with sox32 four-guide Cas9 RNP were collected at 10 hpf and sox17 expression was measured by qPCR. Expression of sox17 mRNA in scrambled control-targeted and sox32-targeted embryos relative to uninjected controls is shown. Graph represents mean ± SEM (n = 4). ∗∗∗p < 0.001, unpaired two-tailed t test. (D) sox17 expression in embryos cell-injected (black) or yolk-injected (gray) with scrambled (scr) or sox32 four-guide Cas9 RNP at the 1-cell stage and collected at 6 or 8 hpf. Expression is normalized to that in uninjected embryos. ∗∗∗∗p < 0.0001; ns, not significant by one-way ANOVA with Sidak's multiple comparisons test compared with equivalent scr experiment in injection method and collection time. (E) Phenotype durability. Embryos were injected with a four-guide Cas9 RNP mix targeting tyr and raised to adulthood. A representative 2-month-old G0 tyr-targeted zebrafish is on the right; control is on the left. Scale bar, 2 mm. Note lack of dark stripes characteristic of tyr knockouts. |
|
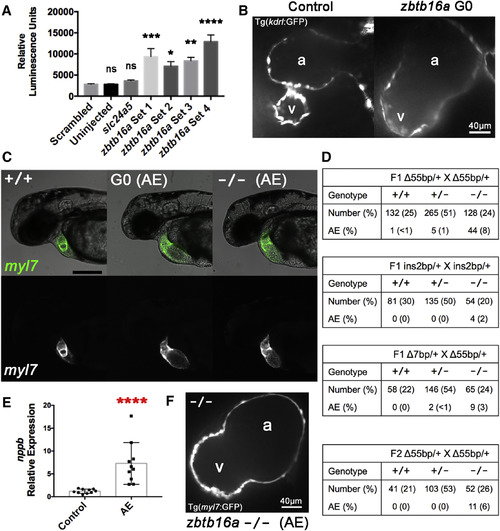
zbtb16a Disruption by Cas9 RNP Impairs Cardiac Development (A) Confirmation of increased nppb expression in response to zbtb16a disruption. Tg(nppb:F-Luc) embryos were yolk-injected with four distinct four-guide sets targeting zbtb16a or with control scrambled and slc24a5 guide sets, then collected at 2 dpf for luciferase assay. zbtb16a guide sets 1–3 targeted the coding sequence as in Figures 6A and 6B. zbtb16a guide set 4 specifically targeted the C2H2 zinc finger repeat domain. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant compared with scrambled by one-way ANOVA and Sidak's multiple comparison test. Mean ± SEM; n = 24 for each group. (B) Representative spinning-disk confocal images of hearts from Tg(kdrl:GFP) embryos collected at 2 dpf after injection with Cas9 RNP with zbtb16a four-guide set (right) or a scrambled control set (left). Tg(kdrl:GFP) labels endocardium here. a, atrium; v, ventricle. (C) Representative images of zbtb16a wild-type Tg(myl7:GFP) embryo (left), Cas9 RNP-injected G0 embryo with atrial enlargement phenotype (AE) (middle), and F3 zbtb16a homozygous null embryo with AE phenotype (right) at 48 hpf. The AE phenotype includes marked chamber enlargement, pericardial edema, and impaired circulation. Fluorescent images at the bottom show myocardium. Scale bar, 250 μm. (D) Genotype and phenotype of F2 and F3 embryos derived from F1 zbtb16a Δ55bp heterozygous incrosses, F1 zbtb16a ins2bp heterozygous incross, F1 zbtb16a Δ7bp heterozygote/Δ55bp heterozygote transcross, and incross of F2 zbtb16a Δ55bp heterozygotes that had been outcrossed once with wild-type. All embryos were scored for AE phenotype at 2 dpf and then genotyped. (E) Quantification of nppb expression in AE phenotype embryos. AE phenotype embryos were selected from zbtb16a Δ55bp heterozygous incrosses and compared with clutchmate controls not exhibiting a cardiac phenotype. ∗∗∗∗ p < 0.0001, unpaired two-tailed t test. (F) Representative spinning-disk confocal image of heart from AE phenotype homozygous zbtb16a Δ55bp null Tg(myl7:GFP) embryo collected at 2 dpf. a, atrium; v, ventricle. PHENOTYPE:
|
|
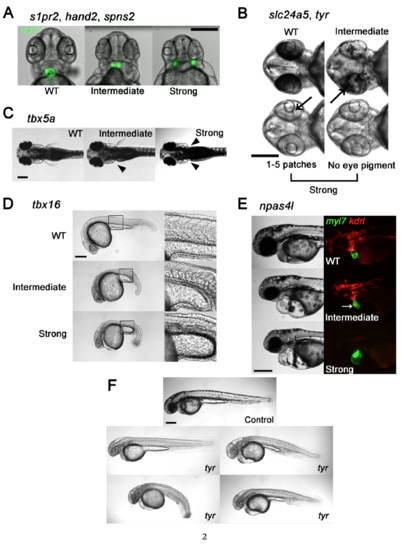
Genes, Phenotypes, and Scoring Conventions. (Related to Figures 1-6) Phenotypes that mimicked those seen in nulls produced by heterozygous mutant lines were scored as Strong and less severe phenotypes were scored as Intermediate as follows: (A) s1pr2, hand2, and spns2. Heart fields marked by Tg(myl7:GFP), showing normal cardiac morphology (WT), partially-fused and non-functional heart (Intermediate), and cardia bifida (Strong). A Strong cardia bifida phenotype was called if the heart fields were completely separated, independent of their distance. (B) slc24a5 and tyr. Black arrows point to patches of pigmented retinal epithelium. No eye pigment or fewer than 5 small patches was classified as a Strong phenotype. Reduced pigmentation but not Strong was classified as Intermediate. (C) tbx5a. Absence of pectoral fin(s) denoted by solid arrowhead(s). Absence of both fins was classified as a Strong phenotype. Absence of one was Intermediate. (D) tbx16. Intact somite formation with abnormal tail bud formation was classified as Intermediate. phenotype. Lack of somite formation and abnormal tail bud formation was classified as Strong phenotype. (E) npas4l. Embryos with cardiac labeling by Tg(myl7:GFP) and endothelial/endocardial labeling by Tg(kdrl:rasCherry) were used. A Strong phenotype was defined as a cloche (bell) heart and absence of endothelial marker. A non-functional misshapen heart (white arrow) with partial endothelial/endocardial labeling was classified as Intermediate. Scale bars, 250 μm. (F) Spectrum of body morphologies in embryos injected with Cas9 RNP with 16 guides directed at the tyr allele. Note embryo with grossly normal body shape as upper left and three dysmorphic embryos with curvature or gross tail, trunk or yolk malformations. Scale bar 250 μm for all images. |
|
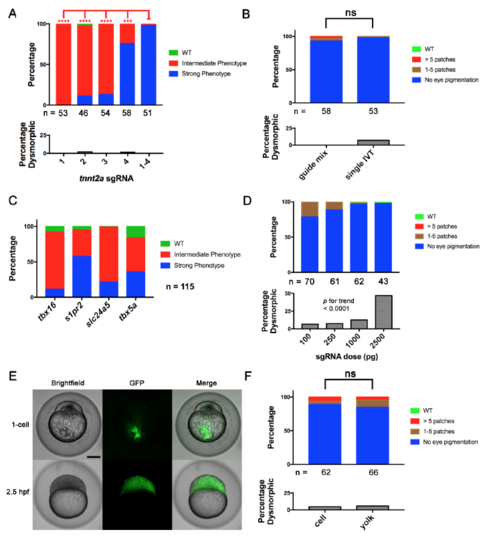
Four-Guide Set Cas9 RNP Generation, Dosing, and Route of Injection. (Related to Figure 2) (A) Penetrance of gene-specific phenotype and toxicity (as % dysmorphic) in embryos after single and four-guide Cas9 RNP injections targeting tnnt2a. **** = p < 0.0001, *** = p < 0.001 Fisher’s exact test for Strong vs. other phenotype (B) A four-guide Cas9 RNP mix for slc24a5 was generated by mixing individually produced sgRNAs (guide mix) or by pooled in vitro template elongation and transcription (single IVT). Cell-injection of either at the one-cell stage produced the slc24a5 depigmentation phenotype and toxicity at a similar rate at 2 dpf. (C) Cas9 RNP containing a pool of 4 sgRNAs targeting 4 different genes produced by a single template elongation and in vitro transcription was cell-injected into 1-cell stage embryos. Embryos were scored for the phenotypes of each individual gene disruption listed at 1 dpf (tbx16, s1pr2), 2 dpf (slc24a5), and 3 dpf (tbx5a). Control injections of a scrambled guide set demonstrated none of the phenotypes. Scoring was performed blinded to treatment. (D) % of embryos with depigmentation phenotype and % dysmorphic at 2 dpf after cell-injection of increasing doses of slc24a5 sgRNA preassembled with 800 pg of Cas9 protein. Increasing dose was associated with increasing toxicity. p < 0.0001 by Chi-square test for trend. (E) Transfer of scrambled sgRNA Cas9-GFP RNP to cells after yolk injection. Brightfield and epifluorescence images were acquired immediately and 2.5 hours after injection. Note complete transfer of fluorescence from injection site in yolk to cells within 2.5h. Scale bar 250 μm. (F) Comparison of yolk vs. cell injection of slc24a5 Cas9 RNP at the one-cell stage on pigmentation at 2 dpf. ns = non-significant for difference in rate of Strong phenotype or dysmorphic phenotype. All experiments were done at least twice with similar results. In A, B and D, the numbers of nondysmorphic embryos analyzed is shown below each column. |
|
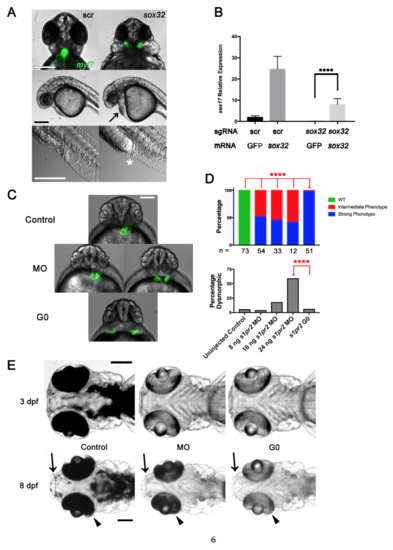
G0 Cas9 RNP Rescue and Comparison of G0 Cas9 RNP to Morpholino Knockdown. (Related to Figure 3) (A) Late sox32 phenotypes. Embryos were cell-injected at the one-cell stage with four-guide Cas9 RNP targeting sox32 or scrambled (scr) control, imaged at 24 hpf. Top panel showing Tg(myl7:GFP) labeling of the heart and cardia bifida after four-guide targeting of sox32. Middle panel showing cardiac edema (black arrow) after four-guide targeting of sox32. Bottom panel showing intestinal and cloacal agenesis (white asterisk). Scale bars 250 μm. (B) sox32 rescue. Embryos were yolk-injected at the one-cell stage with either four-guide Cas9 RNP targeting sox32 or scrambed control and either sox32 or GFP mRNA. Embryos were collected at 6 hpf and sox17 expression was measured by qPCR. **** p < 0.0001 one-way ANOVA with Tukey multiple comparisons test. Mean +/- SEM; n = 4 for each group. (C) Epifluorescence images of uninjected (Control), s1pr2 morpholino (MO), and s1pr2 four-guide Cas9 RNP-injected Tg(myl7:GFP) (G0) embryos at 48 hpf. Representative embryos are shown. About half of the MO-injected embryos showed incomplete fusion of the heart fields, an intermediate phenotype (left) with the remainder showing complete cardia bifida (right) but often less widely spaced than in four-guide Cas9 RNP-targeted embryos (bottom). Scale bar 250 μm. (D) Quantification of the phenotype penetrance and toxicity of embryos generated as in (A). Embryos were injected with MO or Cas9 RNP as indicated. Penetrance of the Strong s1pr2-specific phenotype was higher in Cas9 RNP G0 than in morphants at all MO doses tested. **** = p < 0.0001, Fisher’s exact test. The number of non-dysmorphic embryos scored is shown under each column. Percentage Dysmorphic increased with increasing s1pr2 MO dose (p < 0.0001; Chi-square test for trend). (E) Comparison of the durability of tyr Cas9 RNP (G0) or morpholino (MO) knockdown phenotypes. Representative embryos were analyzed at 3 and 8 dpf are shown. Arrows indicate areas of expected perioral pigmentation and arrowheads indicate retinal pigment epithelium at 8 dpf. Scale bars 250 μm. Note relatively similar levels of depigmentation in morphants and G0 Cas9 RNP embryos at 3 dpf but partial recovery of pigmentation in morphants by 8 dpf. Experiments were performed with yolk injections with doses denoted in Methods. All experiments were repeated at least once. |
|
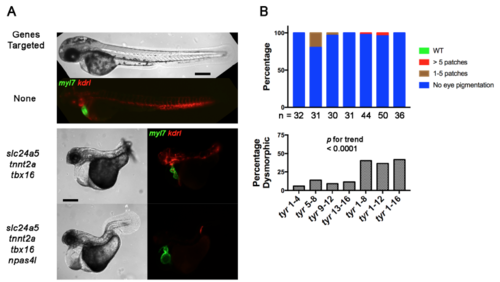
Cas9 RNP Toxicity Increases with sgRNA Number. (Related to Figure 4) (A) Brightfield and epifluorescence images of embryos carrying the Tg(myl7:GFP) cardiomyocyte and Tg(kdrl:rasCherry) endothelial/endocardial markers at 2 dpf. Embryos were left uninjected or cellinjected with multiple four-guide Cas9 RNP mixes targeting slc24a5, tnnt2a, and tbx16, or slc24a5, tnnt2a, tbx16, and npas4l as indicated. Expected phenotypes were present in combination, but the percent of dysmorphic embryos (middle and bottom panels) was high with complex guide sets. Scale bars 250 μm. (B) Percent of embryos with expected phenotype and percent dysmorphic after injection of increasing numbers of guides targeting a single gene. Embryos were yolk-injected with each of four four-guide sets targeting tyr or with these sets in combination such that embryos received 4, 8, 12, or 16 guides. Embryos received the same total amount sgRNA and Cas9 protein independent of guide number. The number of non-dysmorphic embryos analyzed is shown for each condition. Increased guide number was associated with increased toxicity; p < 0.0001 by Chi-square for trend. Scoring was performed blind to treatment. Experiments were performed at least twice with similar results. |
Reprinted from Developmental Cell, 46, Wu, R.S., Lam, I.I., Clay, H., Duong, D.N., Deo, R.C., Coughlin, S.R., A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish, 112-125.e4, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell