- Title
-
A parental requirement for dual-specificity phosphatase 6 in zebrafish
- Authors
- Maurer, J.M., Sagerström, C.G.
- Source
- Full text @ BMC Dev. Biol.
|
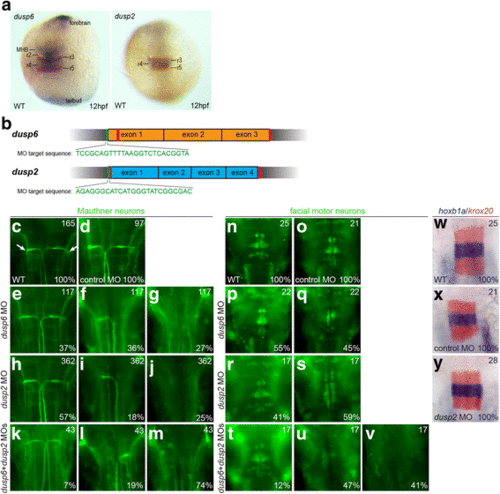
Knockdown of dusp2 and dusp6 via MO yields a hindbrain phenotype. a 12hpf wildtype embryos were assayed by in situ hybridization for expression of krox20 (red stain) and dusp6 (blue stain in left panel) or dusp2 (blue stain in right panel). b Schematic of genomic sequence for dusp6 and dusp2. Red vertical lines indicate CRISPR target sites and green vertical lines indicate MO target sites. c-v 48hpf wildtype (c, n), control MO-injected (d, o), dusp6 MO-injected (e-g, p-q), dusp2 MO-injected (h-j, r-s), and dusp6 + dusp2 MO-injected (k-m, t-v) embryos were assayed by immunostaining for differentiation of Mauthner neurons (3A10 staining in c-m) and facial motor neurons (Islet1/2 staining in n-v). w-y 18hpf wildtype (w), control MO-injected (x) and dusp2 MO-injected (y) embryos were assayed by in situ hybridization for expression of krox20 (red stain) and hoxb1a (blue stain). Numbers in top right corner of each panel indicate the total number of embryos assayed for that condition. Numbers in bottom right corner indicate percent of embryos with the phenotype shown. All embryos are in dorsal view with anterior to the top. Embryos in (a) are whole-mounts, while embryos in (c-y) are flat-mounted and show only the central hindbrain region EXPRESSION / LABELING:
PHENOTYPE:
|
|
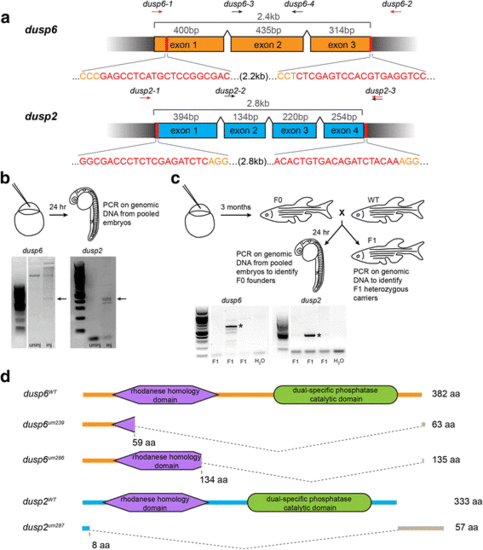
CRISPR genome editing yields loss of function mutants for dusp6 and dusp2. a Schematic of the genomic sequence for dusp6 and dusp2 with the length of each exon and total coding sequence indicated. Black wedges represent introns. Vertical red lines and red nucleotides denote the CRISPR target sequence, and orange nucleotides indicate PAM sequence. Arrows above each schematic indicate the approximate locations of genotyping primers used to detect CRISPR-induced deletion alleles (red arrows) and wildtype alleles (black arrows; see Additional file 2). b Identification of active guide RNAs. Genomic DNA was extracted from pools of injected embryos and PCR-amplified to detect CRISPR-induced deletions. Arrows point to PCR products resulting from successful deletions. c Identification of F0 founder fish. Adult F0 fish were crossed to wildtype and the resulting offspring genotyped as in B. Asterisks indicate transmission of deletions to F1 offspring. d Predicted peptide sequences of the identified mutant alleles for dusp6 and dusp2. The large dashed wedges represent the location of CRISPR-induced deletions, and the gray bars represent residues that are read out of frame prior to a premature stop codon. Amino acid numbers below each peptide sequence indicate the residue affected by the deletion, and the numbers to the right indicate the length of the resulting peptide |
|
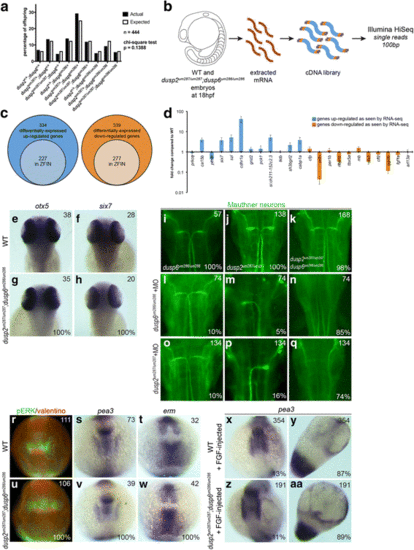
Loss of dusp6 and dusp2 does not impact early development. a Offspring from crosses between double heterozygous dusp2 um287/+ ;dusp6 um286/+ males and females were raised to adulthood and genotyped in order to determine the percentage of each possible genotype in surviving fish. A chi-square test indicates no significant statistical difference between the actual and expected Mendelian ratios. b Outline of RNA-seq library production from wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) 18hpf embryos. c Diagrams representing the number of differentially expressed up-regulated (left circles) and down-regulated (right circles) genes in dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) versus wildtype embryos. Inner circles indicate the subset of genes annotated in ZFIN. d 23 genes identified as differentially expressed by RNA-seq were re-examined by qPCR on cDNA derived from wildtype versus dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos. e-h 48hpf wildtype (e, f) and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males; g, h) embryos were assayed for changes in otx5 (e, g) and six7 (f, h) expression by in situ hybridization. i-q Uninjected (i-k) or MO-injected (l-q) dusp6 mutant (derived from crosses between dusp6 um286/um286 females and dusp6 um286/um286 males; i), dusp2 mutant (derived from crosses between dusp2 um287/um287 females and dusp2 um287/um287 males; j), and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males; k) embryos were assayed by immunostaining with 3A10 antibody to detect the Mauthner neurons at 48hpf. r-w 12hpf wildtype (r, s, t) and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males; u, v, w) embryos were assayed by immunostaining for pERK (green in r, u; red counterstain detects the Valentino transcription factor), as well as by in situ hybridization for expression of pea3 (s, v), and erm (t, w). x-aa Wildtype (x, y) and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males; z, aa) embryos were injected with fgf8 mRNA and the expression pattern of pea3 visualized by in situ hybridization. All embryos are in dorsal view with anterior to the top. Numbers in top right corner of each panel indicate the total number of embryos assayed for that condition. Numbers in bottom right corner indicate percent of embryos with the phenotype shown EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
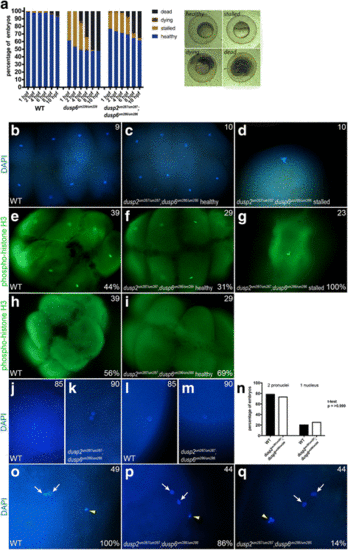
A fraction of dusp6 homozygous mutant embryos stall at the first cell division. a Wildtype, dusp6 mutant (derived from crosses between dusp6 um239/um239 females and dusp6 um239/um239 males), and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) clutches were monitored throughout early development and the health of each embryo scored at each time point according to the brightfield images to the right (a minimum of three clutches were scored for each cross). Average cumulative counts are shown. b-d DAPI staining on 1hpf wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos to visualize the nuclei. e-i Immunostaining for phospho-histone H3 to visualize mitotic nuclei of wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos. j-m DAPI staining at 10 min post fertilization of wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos to visualize the early pronuclei. n Quantification of wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) mutant embryos containing one or two pronuclei. A t-test yields a p-value of > 0.999 indicating no significant statistical difference. o-q DAPI staining to detect polar bodies in wildtype and dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos at 10 min post fertilization. White arrows indicate pronuclei and yellow arrowheads indicate polar bodies in (o-q). Numbers in top right corner of each panel indicate the total number of embryos assayed for that condition. Numbers in bottom right corner indicate percent of embryos with the phenotype shown |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
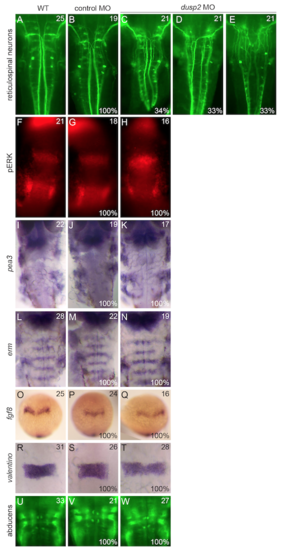
Additional neuronal and patterning markers examined in dusp2 morphants. Wildtype, control MO-injected, and dusp2 MO-injected embryos were analyzed by in situ hybridization for the expression of pea3, erm, fgf8, and valentino and by immunostaining to visualize the reticulospinal neurons, pERK, and the abducens motor neurons. |
|
Additional patterning markers examined in dusp mutants. Wildtype (A, C, E, G, I, K, M, O, Q), dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males; B, D, F, H, J, L, N), dusp2 mutant (derived from crosses between dusp2 um287/um287 females and dusp2 um287/um287 males; P) and dusp6 mutant (derived from crosses between dusp6 um239/um239 females and dusp6 um239/um239 males; R) embryos were analyzed by in situ hybridization for the expression of krox20, fgf3, fgf8, bmp2b, bmp4, chordin, and noggin1. |
|
DAPI staining detects embryos undergoing mitosis. Nuclear staining at approximately 1hpf shows that both wildtype and healthy dusp2/dusp6 double mutant (derived from crosses between dusp2 um287/um287 ;dusp6 um286/um286 females and dusp2 um287/um287 ;dusp6 um286/um286 males) embryos can be detected at stages of mitosis when the chromatids are separated. |