- Title
-
Vegetally localised Vrtn functions as a novel repressor to modulate bmp2b transcription during dorsoventral patterning in zebrafish.
- Authors
- Shao, M., Wang, M., Liu, Y.Y., Ge, Y.W., Zhang, Y.J., Shi, D.L.
- Source
- Full text @ Development
|
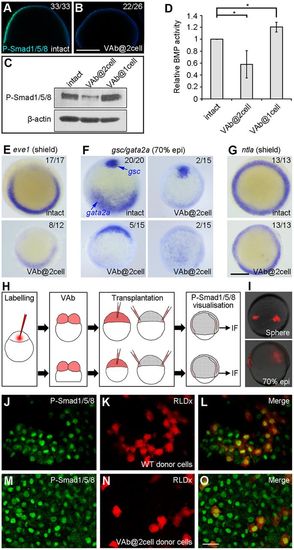
VAb@2cell inhibits BMP signalling. (A,B) Decreased P-Smad1/5/8 immunostaining (green), merged with DAPI (blue), in VAb@2cell embryos at 60% epiboly. (C,D) Analysis by western blotting of P-Smad1/5/8 levels in VAb@2cell and VAb@1cell embryos at 70% epiboly. P-Smad1/5/8 levels are normalised to β-actin, and BMP activity in intact embryos is set as 1. Data are mean±s.d. from three independent experiments. (E-G) Analysis by in situ hybridisation of indicated genes. Animal pole views with dorsal at the top. (H) Procedure of cell transplantation to visualise cell non-autonomous inhibition of BMP signalling in VAb@2cell embryos using P-Smad1/5/8 immunofluorescence (IF) staining. (I) RLDx-labelled donor cells transplanted at different locations of the margin in unlabelled intact embryos at 70% epiboly. (J-O) Similar presence of P-Smad1/5/8 immunostaining in transplanted cells derived from intact (J-L) or VAb@2cell (M-O) embryos. Scale bars: 250 µm in A,B,E-G; 20 µm in J-O. EXPRESSION / LABELING:
PHENOTYPE:
|
|
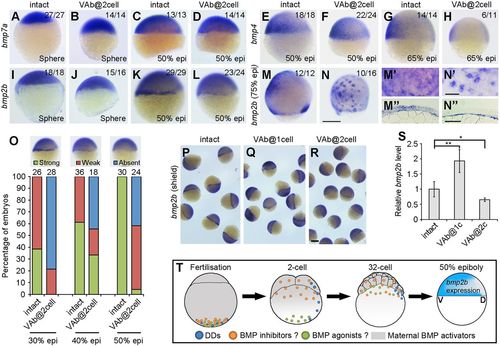
Differential expression of Bmp genes in VAb@2cell embryos. (A-D) Similar expression pattern of bmp7a in intact and VAb@2cell embryos at the indicated stages. (E-H) The expression pattern of bmp4 remains unchanged in VAb@2cell embryos at 50% epiboly, but is strongly reduced at 65% epiboly. (I-L) The initiation of bmp2b expression is unaffected in VAb@2cell embryos at sphere-dome stage, but its zygotic transcription in the margin is reduced or absent at 50% epiboly. (M,N) Scattered bmp2b expression pattern in VAb@2cell embryos at 75% epiboly. Lateral views with dorsal on the right. (M′,N′) Higher magnification in the ventral-animal region of indicated embryos. (M″,N″) Localisation of bmp2b in indicated embryos. (O) Statistical analysis of bmp2b marginal expression in intact and VAb@2cell embryos at the indicated stages, with representative images shown at the top. Numbers on the top of the graph indicate total embryos from two independent experiments. (P-R) Comparison of bmp2b expression in indicated embryos at 50% epiboly. The bmp2b marginal expression is maintained or enhanced in VAb@1cell embryos, but is suppressed in VAb@2cell embryos. (S) Analysis by qRT-PCR of bmp2b levels in VAb@1cell and VAb@2cell embryos. Data are mean±s.d. from four independent experiments. (T) A working hypothesis to explain the dorsalising effect of VAb@2cell. In addition to DDs, putative BMP inhibitors and agonists are also vegetally localised. After fertilisation, the BMP inhibitors should be transported to the animal pole region more rapidly than the BMP agonists; this may account for the appearance of the dorsalised phenotype after VAb@2cell. Scale bars: 250 µm in A-N,P-R; 100 µm in M′-N″. EXPRESSION / LABELING:
PHENOTYPE:
|
|
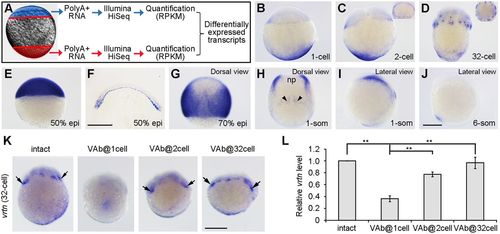
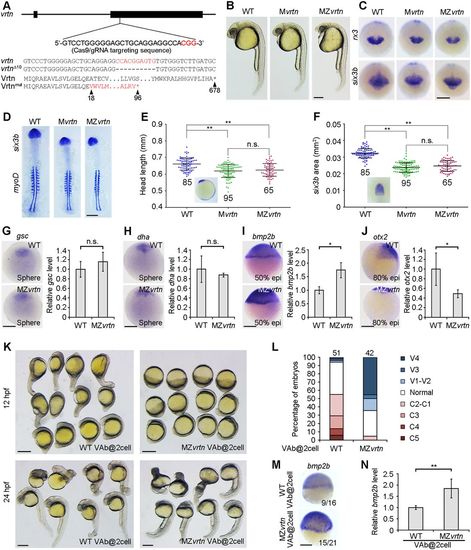
Identification of vegetally localised vrtn transcripts. (A) Procedure for the identification of vegetal pole-enriched transcripts. (B) Maternal vrtn transcripts are localised to the vegetal pole region in the fertilised egg. (C) At the two-cell stage, vrtn transcripts are detected in the marginal region. The inset is an animal pole view. (D) At the 32-cell stage, most vrtn transcripts are localised in the marginal region, as shown by the animal pole view (inset). (E) Strong vrtn expression in the blastoderm at 50% epiboly. (F) Vertical section showing marginal and YSL expression of vrtn at 50% epiboly. (G) Dorsal view at 70% epiboly shows strong vrtn expression in the lateral region. (H,I) Dorsal and lateral views show vrtn expression in the neural plate boundary (np) and forming somites (arrowheads) at the one-somite stage. (J) Lateral view shows vrtn expression in the tail-bud at the six-somite stage. (K) Analysis by in situ hybridisation of vrtn expression following VAb at indicated stages. Arrows indicate the marginal accumulation of maternal vrtn transcripts. (L) Analysis by qRT-PCR of vrtn levels. The relative vrtn level in intact embryos is normalised as 1; data are mean±s.d. from three independent experiments. Scale bars: 250 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
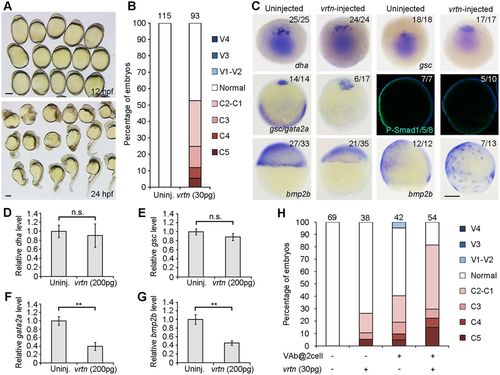
Vrtn overexpression produces dorsalisation and suppresses BMP signalling. (A) Dorsalised embryos at 12 and 24 hpf following Vrtn overexpression. (B) Statistical analysis of Vrtn-dorsalised embryos at 24 hpf. Numbers at the top indicate the total embryos from three independent experiments. (C) Early effects of Vrtn overexpression. The expression patterns of dha/boz and gsc remain unchanged at the sphere stage, and gata2a expression at 70% epiboly and P-Smad1/5/8 level at 60% epiboly are suppressed. The marginal transcription of bmp2b is inhibited at 50% epiboly, and scattered I-YSL expression of bmp2b is present at 70% epibloy. (D-G) Analysis by qRT-PCR of the expression level of dha/boz, gsc, gata2a and bmp2b in Vrtn-overexpressing embryos. The relative expression level in uninjected embryos is normalised as 1; data are mean values±s.d. from four independent experiments. (H) Statistical analysis of the synergistic effect between VAb@2cell and Vrtn overexpression. Numbers at the top indicate the total number of embryos from two independent experiments. Scale bars: 250 µm. |
|
The dorsalising effect of VAb@2cell is suppressed in MZvrtn mutants. (A) Targeted mutation of the vrtn gene. The Cas9/gRNA targeting sequence and the PAM region (red) are shown, and the mutated allele with the predicted peptide is indicated. (B) Representative images of indicated embryos at 27 hpf, showing smaller eyes and reduced head size in vrtn mutants. (C) Decreased rx3 and six3b expression domains in vrtn mutants at the bud stage. (D) Expression patterns of six3b and myod1 in flat-mounted embryos with equal somite numbers. (E,F) Analysis by scatter plots of head length and six3b expression area at the six-somite stage. The red broken lines in the insets define the parameters of measurement. Horizontal lines represent the mean±s.d. from three batches of embryos with the total number indicated. (G-J) Analyses by in situ hybridisation and qRT-PCR of the expression of gsc, dha/boz, bmp2b and otx2 in MZvrtn mutants. The expression of gsc and dha/boz remains unchanged at sphere stage (animal pole views), the expression of bmp2b is enhanced at 50% epiboly, and the expression of otx2 is reduced at 80% epiboly (lateral views with dorsal on the right). The relative expression level in wild-type embryos is normalised as 1; data are mean±s.d. from three independent experiments. (K) MZvrtn embryos are ventralised following VAb@2cell. (L) Statistical analysis of the ventralising effect. Numbers at the top represent total embryos from three independent experiments. (M,N) Analyses by in situ hybridisation and qRT-PCR of bmp2b expression at the shield stage in wild-type and MZvrtn embryos after VAb@2cell. The embryos are shown as lateral views. The relative bmp2b level in VAb@2cell wild-type embryos is set as 1; data are mean±s.d. from six independent experiments. Scale bars: 250 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
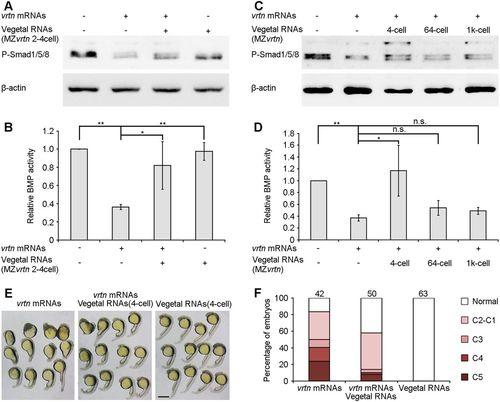
Vegetal RNAs antagonise the dorsalising and BMP-inhibiting activity of Vrtn. (A) Western blotting at 50% epiboly shows the rescue of P-Smad1/5/8 levels in Vrtn-overexpressing embryos by vegetal RNAs from the two- to four-cell stage. (B) P-Smad1/5/8 level is normalised to β-actin and quantified with respect to uninjected embryos. Data are mean±s.d. from four independent experiments. (C) Western blotting shows the temporal regulation of Vrtn activity by vegetal RNAs. (D) Quantification of P-Smad1/5/8 level. Vegetal RNAs from 64- or 1000-cell stages show no effect on Vrtn-inhibited P-Smad1/5/8 levels. Data are mean±s.d. from three independent experiments. (E,F) Representative embryos at 24 hpf and statistical analysis show that vegetal RNAs counterbalance Vrtn-mediated dorsalisation. Numbers at the top indicate total embryos from two independent experiments. Scale bar: 200 µm. PHENOTYPE:
|
|
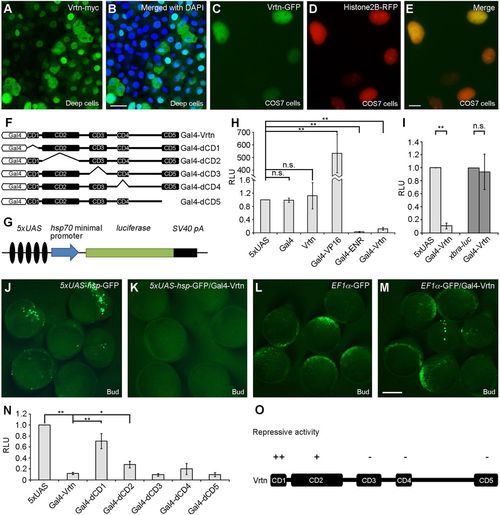
Vrtn functions as a transcriptional repressor. (A,B) Nuclear localisation of Vrtn-myc in deep cells of zebrafish embryos at 50% epiboly. (C-E) Nuclear localisation of Vrtn-GFP in COS7 cells. (F) Schematic of full-length and truncated Vrtn fused with a Gal4 DNA-binding domain. (G) Schematic shows the 5×UAS-hsp-luc reporter. (H) Graph shows the specificity and validity of the one-hybrid assay system. Data are mean±s.d. from three independent experiments. (I) Gal4-Vrtn has no effect on the xbra promoter. Data are mean±s.d. from three independent experiments. (J-M) Gal4-Vrtn suppresses GFP transcription driven by the 5×UAS-hsp promoter, but not by the EF1α promoter. The embryos were injected with 5×UAS-hsp-GFP plasmid (100 pg) or EF1α-GFP plasmid (20 pg), either alone or together with Gal4-Vrtn mRNA (200 pg). (N) Graph shows the transcriptional repressor activity of full-length and truncated Vrtn. Data are mean±s.d. from three independent experiments. (O) Summary of the repressive activity of Vrtn domains. Scale bars: 20 μm in A,B; 10 µm in C-E; 200 µm in J-M. |
|
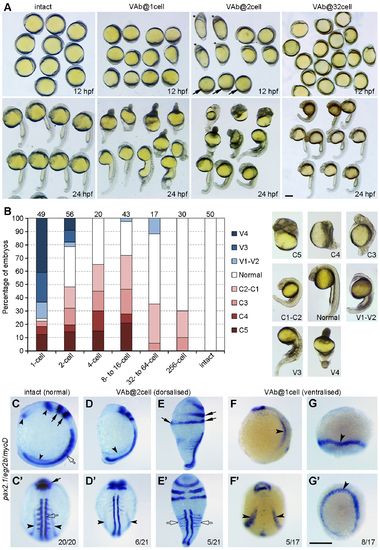
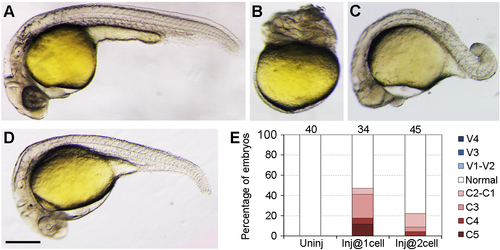
Stage-dependent dorsalising effect of VAb. (A) Representative phenotypes following VAb at indicated stages. Asterisks and arrows denote dorsalised and ventralised embryos, respectively. (B) Bar graph shows the stage-dependent dorsalising effect of VAb. Numbers on the top indicate total embryos scored from three independent experiments. Different degrees of dorsalised (C5-C1) and ventralised (V4-V1) phenotypes are shown on the right. (C-G') Dorsalised and ventralised embryos at 13 hpf revealed by simultaneous ISH. pax2.1 labels optic field, midbrain-hindbrain boundary and pronephros (black arrowheads), egr2b (krox20) labels the hindbrain (black arrows), and myoD marks the somites (open arrows). The images are shown in lateral (C,D,E,F,G) and dorsal (C',D',E',F',G') views, with anterior on the top. (C,C') An intact embryo. (D,D') A mildly dorsalised VAb@2cell embryo. (E,E') A severely dorsalised VAb@2cell embryo with radial neural marker expression and expanded paraxial tissue. (F,F') A mildly ventralised VAb@1cell embryo. (G,G') A radially ventralised VAb@1cell embryo with only pronephric progenitor expression of pax2.1. Scale bars: 250 μm. |
|
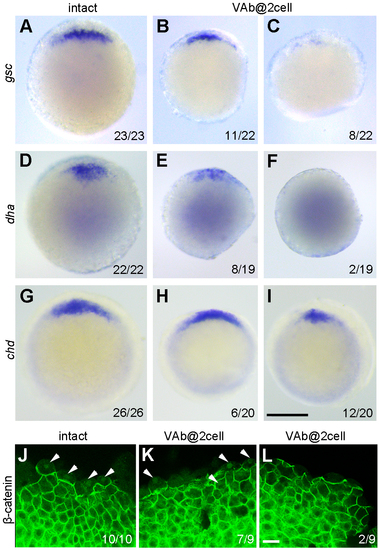
The activation of maternal Wnt/ß-catenin signalling is either unaffected or reduced after VAb@2cell. (A-I) Analysis by ISH of gsc, dha, and chd expression at sphere-dome (A-F) and 50% epiboly (G-I) stages. Animal pole views with dorsal region on the top. (J-L) Immunofluorescence staining of ß-catenin nuclear localisation (arrowheads) in intact and VAb@2cell embryos at 3.5 hpf. Scale bars: (A-I) 250 μm; (J-L) 20 μm. |
|
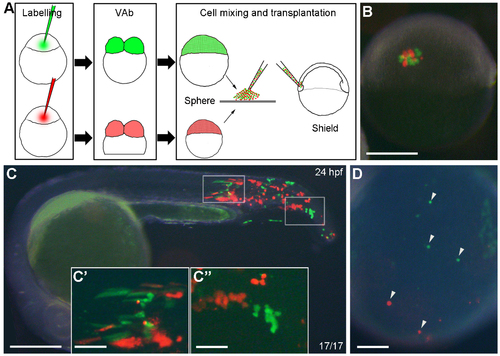
Transplanted cells from VAb@2cell embryos can differentiate into ventral tissues. (A) Diagram illustrating the procedure of cell transplantation to compare the distribution of descendent cells from intact (green) and VAb@2cell (red) embryos in unlabelled intact recipients. (B) A successfully transplanted embryo at shield stage. (C-C'') Comparable localisation of green and red cells in a chimeric embryo at 24 hpf, with higher magnifications of the boxed regions. (D) Transplanted cells from intact and VAb@2cell embryos can similarly differentiate into circulating blood cells (arrowheads) in the yolk sac circulation valley of a 24 hpf chimera. Scale bars: (B,C) 250 μm; (C'-D) 50 μm. |
|
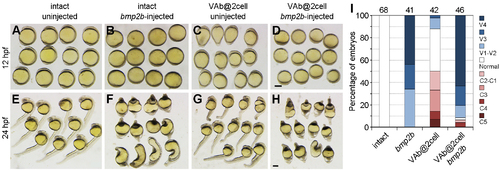
The dorsalising effect of VAb@2cell is reversed by bmp2b overexpression. (A-H) Representative phenotypes from indicated embryos at 12 hpf and 24 hpf. (I) Statistical analysis of different phenotypes. Numbers on the top indicate total embryos scored from two independent experiments. Scale bars: 250 μm. |
|
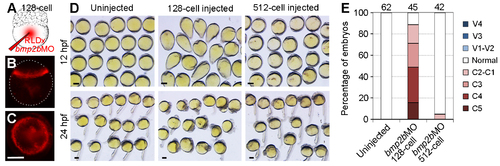
Knockdown of bmp2b in the margin mimics the dorsalising effect of VAb@2cell. (A) Diagram showing the coinjection of bmp2bMO (2 ng/embryo) and RLDx in the yolk cell at 128-cell stage. (B,C) RLDx labelling in the margin of a sphere stage embryo with lateral (B) and animal pole (C) views. White dots outline the embryo. (D) Representative phenotypes at 12 hpf and 24 hpf resulted from bmp2b knockdown at indicated stages. The distribution of bmp2bMO is only restricted in YSL following injection in the yolk cell at 512-cell stage. (E) Comparison of the efficiency of dorsalisation following bmp2b knockdown at 128-cell and 521-cell stages. Numbers on the top indicate total embryos scored from two independent experiments. Scale bars: 250 μm. |
|
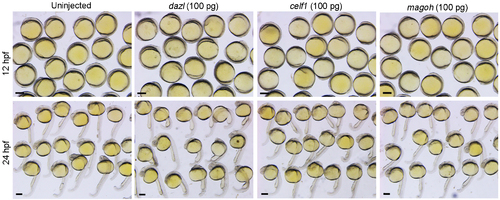
Absence of dorsalising effect following overexpression of dazl, celf1 or magoh. Representative phenotypes of embryos at 12 hpf and 24 hpf previously injected with indicated mRNAs. Compared to uninjected embryos, overexpression of the three genes does not affect the formation of DV axis. Scale bars: 250 μm. |
|
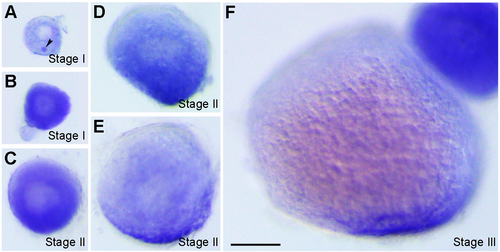
Localisation of vrtn transcripts during oogenesis. (A) Stage I oocyte (80 μm in diameter), with vrtn transcripts detected in the ooplasm, and in the Balbiani body (arrowhead). (B) Stage I oocyte (100 μm in diameter), with strong uniform localisation in the ooplasm. (C) Stage II oocyte (150 μm in diameter) shows a slight vegetal enrichment. (D) Stage II oocyte (200 μm in diameter), with evident vegetal localisation. (E) Stage II oocyte (230 μm in diameter) shows vrtn localisation mainly in the vegetal region. (F) Stage III oocyte with vrtn transcripts restricted exclusively in the vegetal pole cortex. Oocytes were staged according to their diameters: stage I, 70-140 μm; stage II, 140-340 μm; stage III, 340-690 μm. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Stage-dependent dorsalisation following Vrtn overexpression. (A) An uninjected embryo at 24 hpf. (B-D) Different degrees of dorsalised embryos, which can be categorised into C5 (B), C3 (C) and C2 (D), respectively. (E) Stage-dependent decline of the dorsalising effect following Vrtn overexpression. Notice the weak dorsalising activity when vrtn mRNA is injected after first cleavage stage. Numbers on the top indicate total embryos scored from two independent injection experiments. Scale bar: 250 μm. |
|
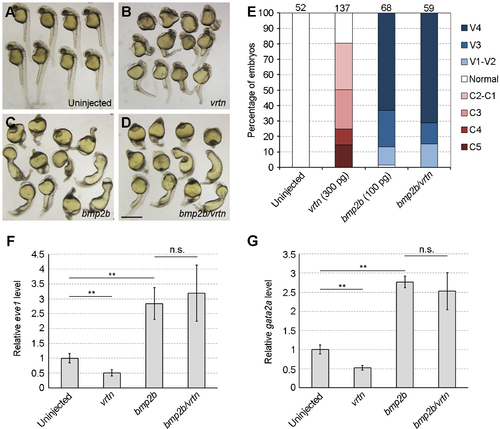
Vrtn does not affect signal transduction by Bmp2b. (A-D) Phenotype analysis of indicated embryos at 28 hpf. The ventralised phenotype resulted from Bmp2b overexpression is not reversed by the presence of Vrtn. (E) Statistical analysis of the phenotypes. Numbers on the top indicate total embryos scored from two independent experiments. (F,G) Analysis by qRT-PCR of eve1 and gata2a expression at 75% epiboly. The relative expression level of eve1 and gata2a in uninjected embryos is normalised as 1, and bars represent the mean values ± s.d. from three independent experiments. Vrtn does not affect eve1 and gata2a expression induced by Bmp2b. Scale bar: 800 μm. |
|
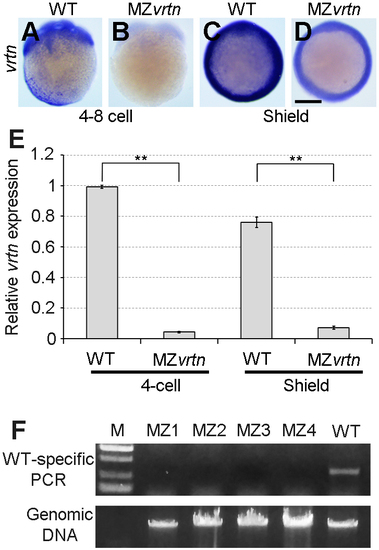
Validating the genotype of MZvrtn embryos. (A-D) Analysis by ISH of maternal and zygotic vrtn transcripts in WT and MZvrtn embryos at 4- to 8-cell (lateral view) and shield (animal pole view) stages. (E) Analysis by qRT-PCR shows the non-sense mediated decay of vrtn mutant mRNAs in MZvrtn embryos. The value of vrtn transcripts in 4-cell stage WT embryos is set as 1, and the expression level is calculated as the ratio between vrtn and ß-actin. Bars represent the mean values ± s.d. from three independent experiments. (F) Genotyping of 4 MZvrtn embryos (MZ1 to MZ4) at 24 hpf, using a WT-specific PCR primer. A WT embryo is used as a positive control. The input genomic DNA serves as a loading control. |
|
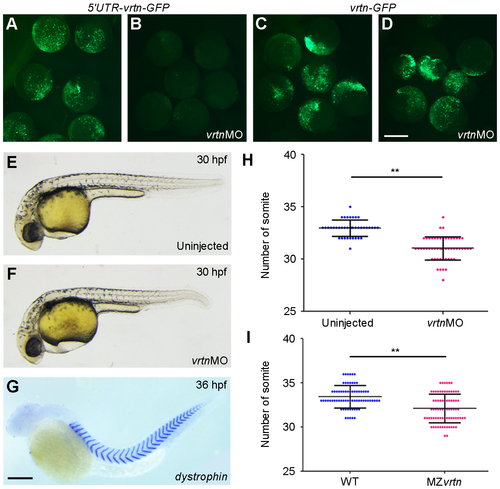
Somite number is reduced in vrtn morphants or mutants. (A-D) Specificity of the vrtnMO. Injection of vrtnMO efficiently blocked the translation of the artificial mRNA where the vrtnMO-targeting 5' leader sequence is fused with vrtn-GFP. It has no effect on the artificial mRNA where only vrtn coding region is fused with GFP sequence. (E,F) Phenotypes of uninjected and vrtnMO-injected embryos at 30 hpf. (G) Analysis of the number of somite boundary at 36 hpf after ISH using dystrophin as a marker. (H,I) Statistical analysis of somite number in vrtn morphants and mutants. Horizontal lines represent the mean values ± s.d. from four batches of embryos. Scale bars: (A-D) 400 μm; (E-G) 250 μm. PHENOTYPE:
|
|
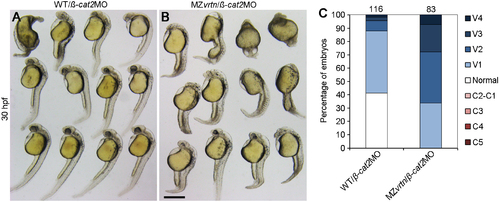
MZvrtn embryos are more sensitive to ventralisation following ß-catenin2 knockdown. (A,B) Representative phenotypes of WT and MZvrtn embryos injected with ßcat2MO (5 ng). Note the increase in the number of embryos with absence of eye in ß-cat2MO injected MZvrtn mutants. (C) Statistical analysis of ß-catenin2 knockdown in WT and MZvrtn embryos. Numbers on the top indicate total embryos counted in three independent experiments. Scale bar: 200 μm. |
|
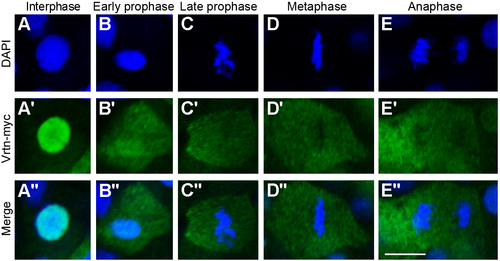
Nuclear localisation of Vrtn during interphase of the cell cycle. Immuofluorescence staining of Vrtn-myc during different phases of the cell cycle in zebrafish embryos at 50% epiboly. (A-E) DAPI staining of nuclear DNA. (A'-E') Immunofluorescence staining of Vrtn-myc. Nuclear localisation is detected only during the interphase. (A''-E'') Merge of DAPI and Vrtn-myc staining. Scale bar: 10 μm. |
|
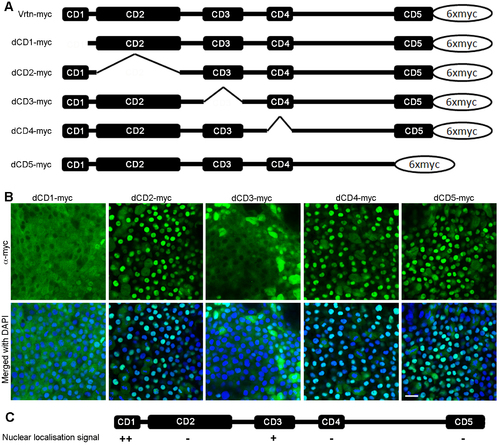
Subcellular localisation of Vrtn deletion mutants. (A) Diagram showing full-length and truncated forms of Vrtn fused with 6 myc epitopes at the C-terminus. (B) Confocal images showing the distribution of Vrtn deletion mutants in deep cells of embryos at 50% epiboly stage. Note that the nuclear localisation of Vrtn mutants lacking CD1 (dCD1) or CD3 (dCD3) is obviously reduced. (C) Summary of the nuclear localisation activity of different Vrtn deletion mutants. Scale bar: 20 μm. |