- Title
-
An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits
- Authors
- Förster, D., Dal Maschio, M., Laurell, E., Baier, H.
- Source
- Full text @ Nat. Commun.
|
Optobow allows all-optical mapping of connected neurons. a Design of the Optobow-c construct. mCerulean and tdTomato are both membrane-targeted (CAAX motif). b Sketch for Optobow-c expression in the zebrafish optic tectum. Different cell types are indicated by their arborization patterns in the tectal neuropil. Addition of Cre by transient injections results in random expression of either ChrimsonR (labeled by tdTomato, magenta) or GCaMP6f (green). Unrecombined cells are labeled by mCerulean (blue). Upon photostimulation of a single ChrimsonR cell (dashed orange outline) connected cells will be highlighted by a rise in GCaMP fluorescence (dark green cell). c In vivo tectal expression of Optobow-c. Scale bar, 20 µm. d Microscope setup for all-optical connectivity mapping. Two independent infrared-pulsed lasers are used to photostimulate ChrimsonR cells (magenta) and to image GCaMP fluorescence (green), respectively. A spatial light modulator (SLM) allows precisely targeted stimulation of single cells. See methods for abbreviations. e Dorsal view of tectal Optobow-c expression. Single fluorescent channels show a cluster of three ChrimsonR- (left) and 18 GCaMP6f-expressing cells (middle). Photostimulation was either confined to a single ChrimsonR cell (Stim1, dashed orange outline) or to a control region of equal size (Stim2). White dashed line marks skin. Dotted rectangles show regions of calcium imaging during photostimulation. A close-up showing a single confocal plane (z = 2.5 µm) of the photostimulated region (Stim1) is shown in the merge on the right. Despite the close proximity of the ChrimsonR and the GCaMP cell#2, exclusive expression of either component allows restricted stimulation of the ChrimsonR-expressing cell. Scale bar, 15 µm. f Calcium transients acquired at 10 fps shown as Z-scores, obtained simultaneously from the regions indicated in e. Photostimulation events are highlighted by dashed lines with stimulation lengths indicated below. Off-target stimulation did not result in significant GCaMP activity (Stim2) |
|
Simultaneous connectivity mapping and morphological analysis with Optobow-n. a Schematic of the Optobow-n construct. b Tectal-specific expression of Optobow-n. A single ChrimsonR-expressing cell (magenta) is surrounded by seven nls-GCaMP6f-expressing cells, membrane-labeled by mCitrine (yellow). Localization of GCaMP6f (cyan) appears completely restricted to the nucleus. White dashed line indicates skin. Orange dashed line in two-channel merge (right image) marks photostimulated region. Dotted rectangles show regions of calcium imaging during photostimulation. Arrowheads show mCerulean signals in non-recombined cells. Scale bar, 20 µm. c Calcium transients (Z-scores) acquired at 10 fps from the regions annotated in b. Photostimulation events of 200 ms are marked by dashed lines. While calcium activity of cell#1 is tightly coupled to the photostimulation, activity of cell#2 appears slightly delayed. No significant calcium responses were detectable in other neighbouring cells (#3–5). d Three-dimensional tracings of the photostimulated cell (magenta) and the two responding cells. Dorsal view is shown on the left and transverse view on the right. The stimulated cell is a bistratified projection neuron with a descending axon (arrowhead). Cell#1 is a bistratified periventricular interneuron and cell#2 is a non-stratified projection neuron, which sends an axon to the intertectal commissure (arrows). Scale bar, 20 µm |
|
PA-GFP reveals morphologies of functionally connected cells. a In Optobow-nPA, all ‘nls-GCaMP6s’ cells co-express PA-GFP. b Optobow-nPA expression in the optic tectum. A single ChrimsonR-expressing cell (magenta) was photostimulated (orange dashed line) and GCaMP fluorescence was monitored in six neighbouring cells by line scans across the nuclei. Note that PA-GFP is not detectable before its activation. Scale bar, 20 µm. c Calcium measurements acquired at 250 fps for the GCaMP cells annotated in b. Raw (grey) and averaged ∆F/F responses (red) are shown on the left, heat maps for Z-scores are on the right. Photostimulation epochs of 200 ms are indicated by dashed lines. Cell#1 and #5 showed reliable calcium responses upon photostimulation. d Close-up of cell#1 after photoactivation of PA-GFP. A less saturated, single slice of the cell body region shown in the lower left demonstrates exclusive photoactivation of cell#1. Scale bar, 20 µm. e, f Three-dimensional filament reconstruction of the presynaptic cell (magenta) and cell#1 (green) in dorsal view (e) and transverse view (f). The presynaptic cell is a bistratified projection neuron, and cell#1 is a non-stratified projection neuron. Descending projection axons are marked (arrows). Scale bar, 20 µm |
|
Optobow-nPA_Syp reveals potential synaptic contacts. a In Optobow-nPA_Syp, ChrimsonR-expressing cells are co-labeled by Synaptophysin-mCitrine. b Tectal-specific expression of Optobow-nPA_Syp. Cell bodies of two ChrimsonR-expressing cells (magenta, close-up in lower right, Stim1) or neuropil regions (Stim2), respectively, were stimulated (orange dashed line), and activity of two nls-GCaMP6s-expressing cells (arrowheads) was monitored. Note that a radial glia cell overexpressing ChrimsonR-tagRFP and Syp-mCitrine appears in white. c Z-scores of calcium measurements for cell#1 and cell#2 during cell body (Stim1) or neuropil stimulations (Stim2). Photostimulation epochs of 200 ms are indicated by dashed lines. Cell#1 shows high response reliability. While cell#2 shows spontaneous activity during Stim1, its response correlates to neuropil stimulations suggesting that additional ChrimsonR-labeled cells were activated by Stim2. d Photoactivation of PA-GFP in cell#1 (arrow). Spectral unmixing was used to separate PA-GFP/GCaMP (green) from mCitrine signals (yellow; see Methods section). Scale bar, 20 µm. e Close-up on single confocal slices of the regions marked in d before and after PA-GFP photoactivation. Scale bar, 10 µm. f Three-dimensional filament reconstruction of presynaptic (magenta) and connected cells (green) in transverse view. Potential synapses, shown in yellow (arrows), are restricted to a single tectal layer. Scale bar, 20 µm |
|
Quantification of response latencies and reliabilities. All data are derived from 5 dpf larvae expressing Optobow-nPA, using a sampling rate for calcium signals of <4.5 ms per frame. a Distribution of all response latencies. Events are grouped in temporal bins of 50 ms. Orange shaded region marks 200 ms of ChrimsonR photostimulation. Number of trials n = 65, number of cells = 12. b Individual cell response reliability vs. response onset latency. For every cell, the average response onset is represented along the X axis, while the ratio responses per trials is plotted in the Y axis. Dashed line represents a linear regression model of the data to evaluate the degree of correlation (coefficient of determination is 0.13329). Morphological analysis shown below indicates overlapping (a–c) or non-overlapping (d–f) neurite arbors of the functionally identified cell pairs. Error bars are SD. Scale bar, 20 µm. c Average response latencies of three non-overlapping cells (red data points in b) compared to all other identified responding cells. Error bars are SD |
|
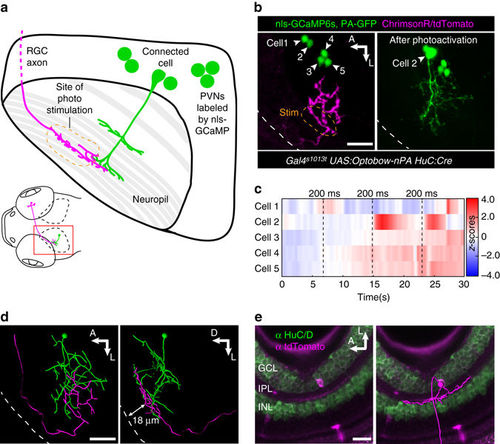
Optobow analysis can be expanded to study long-range connectivity of different cell types. a Sketch simplifying the connection of an RGC axon with a PVN in the optic tectum. b Live expression of Optobow-nPA. A single RGC axon (magenta) and five PVN nuclei are labeled (green, left). White dashed lines mark the skin. Upon identification of the connected cell#2, PA-GFP was photoactivated (right). Scale bar, 20 µm. c Calcium traces (Z-scores) for the cells annotated in b. Photostimulation epochs of 200 ms are indicated by dashed lines. Only cell#2 showed responses upon photostimulation. d Filament reconstructions in dorsal view (left) and transverse view (right). The RGC axon terminates in the stratum fibrosum et griseum superficiale (SFGS) layer 1 (18–20 µm distance from skin), where it contacts the dendrites of the bistratified PVIN (cell#2). Scale bar, 20 µm. e Immunostainings against HuC/D (green) and tdTomato (magenta) reveal the dendritic pattern of the activated RGC in the retina. A filament reconstruction is shown on the right. Its monostratified dendrites exclusively target the OFF layer of the IPL. Scale bar, 20 µm. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; PVIN, periventricular interneuron |
|
Optobow reveals novel excitatory connections of projection neurons within the optic tectum. a, b Morphological reconstructions of connected tectal cells, identified using Optobow-nPA, in dorsal view (a) and transverse view (b). Cell types are indicated on the left. White dashed lines mark the skin. Arrows point to projection axons leaving the neuropil. Values for response onset time (s) and response reliability (r) are shown in b. Scale bar, 15 µm. c Model for tectal connectivity of PVPNs. In addition to an axon leaving the tectum, PVPNs make functional, excitatory connections both with PVINs and other PVPNs. bs, bistratified; ms, monostratified; ns, non-stratified; PVIN, periventricular interneuron; PVPN, periventricular projection neuron; ts, tristratified |
|
Registration of Optobow fish highlights relative anatomical positions of connected cell pairs. a–d Registration of six identified cell pairs (illustrated by the same cell body color) into one reference brain (outlined by DAPI). Shown are dorsal (a), lateral (b and c) and frontal views (d). Optic tecta are outlined by white, and eyes by yellow dashed lines, respectively. The RGC-PVIN cell pair (white cell bodies) described in Fig. 6 was added in b–d. Scale bar, 50 µm. e, f Dorsal (e) and transverse views (f) of single cell pairs shown in a–d. White dashed lines outline skin. Identified cell types are indicated below. Scale bar, 20 µm |
|
Stimulation of ChrimsonR-expressing cells elicits behavior in larval zebrafish. (A) Transgenic ChrimsonR-tagRFP expression in the nucleus of the medial longitudinal fasciculus (nMLF) of a 5 dpf zebrafish larva. Scale bar, 20 μm. (B) Optogenetic setup as published previously1. The head of the same fish from (A) is embedded in agarose with its tail freed. A 50 μm optic fiber targets 638 nm light (0.1 mW) onto the region of the nMLF and is moved in a vertical direction. A projection of the initial and maximum left deflected tail positions is shown. Scale bar, 1 mm. (C) Measurement of the tail angle and the fiber position over time of the fish shown in (B). The red rectangle depicts the epoch of 638 nm light stimulation. |
|
List of available Optobow zebrafish lines. Variegation of the different UAS-transgenic lines was scored based on expression of mCerulean-CAAX from the pan-neuronal driver Gal4s1101t. Scale bar: 40 μm. |
|
Characterization of 2P holography photostimulation. (A) Axial (top) and lateral (bottom) extension of a circular illumination pattern with a diameter of 6 μm. 960 nm light was used to excite a thin layer of Fluorescein solution (0.2 mW μm-2). Axial and lateral intensity profiles are shown on the right. Scale bar, 5 μm. (B) Maximum calcium responses obtained upon photostimulation of ChrimsonR-expressing cells using different wavelengths. Excitation of Chrimson at 760 nm resulted in relatively large responses, likely driven by a single photon absorption mechanism. Data obtained from Optobow-nPA experiments in tectal cells. Error bars indicate SEM. (C-E) Characterization of ChrimsonR photostimulation selectivity in vivo. (C) Maximum projection of a confocal stack showing tectal cells in a 6 dpf zebrafish larva with a dense expression of ChrimsonR-tagRFP (left: magenta, right: white) and pan-neuronal nls-GCaMP6s (green). A close-up single confocal slice of the targeted cell is shown below the projection. A 6 μm-diameter excitation spot was used for on-target (orange circle, #1) or off-target (cyan circles, #2-6) photostimulations at 1020 nm (200 ms), while calcium responses of cell#1 were recorded simultaneously. Scale bar, 10 μm. (D) Averaged maximum ΔF/F calcium responses obtained from on-target and off target stimulations as a function of distance between the recorded cell (#1) and the excitation spot. Error bars are SEM. (E) Raw (gray) and averaged (red) ΔF/F profiles of six excitation trials for the different spot positions indicated in (C). Stimulation time points are indicated by arrowheads. |
|
Comparison of cytoplasmic and nuclear GCaMP6 variants. (A) GCaMP6 dynamics were measured in nMLF cells (Gal4s1171t) transiently expressing ChrimsonR-tagRFP and either of the GCaMP6 variants. Single cells were photostimulated with 200 ms of 760 nm light and the GCaMP transients were measured by ~300 Hz line scans across the soma of the stimulated cell. Single-channel closeups shown on the right of the regions indicated by dashed boxes, show membrane, cytoplasmic, or nuclear localization of ChrimsonR-tagRFP, GCaMP6s or nls-GCaMP6s, respectively. G6s, GCaMP6s; G6f, GCaMP6f. Scale bar, 30 μm. (B) Normalized mathematical fit for the fluorescence responses of all measured GCaMP6 versions. Number of trials (n) is indicated in (C) and (D). (C) Comparison of fluorescence rise times to peak intensity. (D) Comparison of half decay times. Error bars indicate SEM. |
|
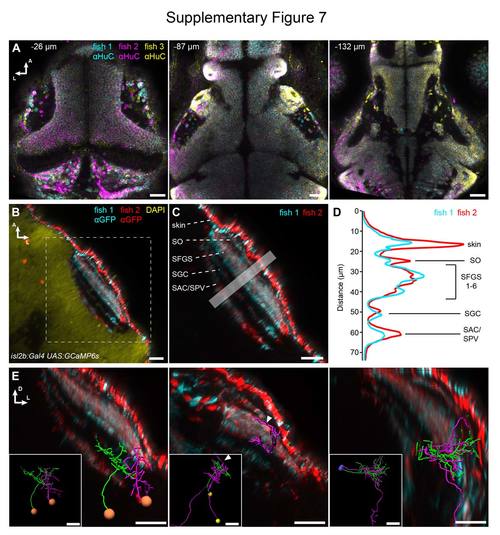
Accuracy of registration and addition of anatomical reference patterns. (A) Three 5 dpf Optobow-nPA-expressing larvae stained for HuC have been registered into one reference brain. Note the accuracy of registration at different z levels (labeled as distance from dorsal skin in μm). Scale bar, 30 μm. (B) Two 5 dpf larvae expressing GCaMP6s under control of isl2b:Gal4 (stained for GFP) were co-registered into the reference brain. Scale bar, 20 μm. (C) A closeup of the tectal neuropil shows the different innervation strata of RGC axons. SO, stratum opticum; SFGS, stratum fibrosum et griseum superficiale; SGC, stratum griseum centrale; SAC/SPV, stratum album centrale/stratum periventriculare. Scale bar, 20 μm. (D) Intensity profiles through the boxed region in (C) show the alignment accuracy of RGC innervation strata. (E) Examples of three co-registered cell pairs and their dendritic and axonal arborizations in specific tectal layers. Insets taken from Figure 8. Scale bar, 20 μm. |