- Title
-
CRISPR/Cas9-Mediated Zebrafish Knock-in as a Novel Strategy to Study Midbrain-Hindbrain Boundary Development
- Authors
- Kesavan, G., Chekuru, A., Machate, A., Brand, M.
- Source
- Full text @ Front. Neuroanat.
|
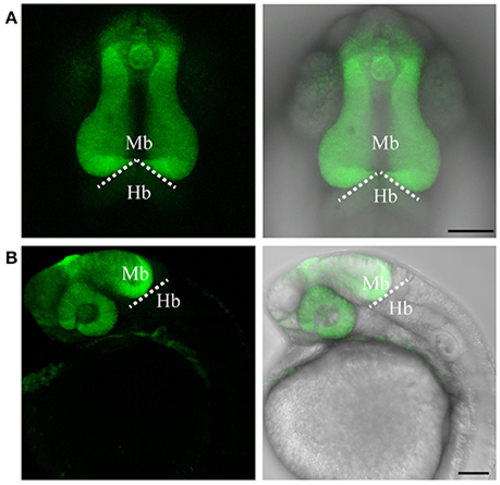
Targeted knock-in of Venus fluorescent protein into the otx2 locus. Expression of Venus fluorescent protein at 24 hpf. Images were taken from live embryos anesthetized in MS-222. (A) Left panel shows dorsal view with the anterior of the embryo facing upwards, right panel is a merged image of the fluorescent and transmitted light channels. Venus is expressed in the retina and midbrain, sharply abutting the MHB (dotted line). (B) Lateral view of an embryo expressing Venus. Left panel shows fluorescent channel and right panel shows a merged image of the fluorescent and transmitted light channels. All images are maximum intensity projections covering 50 μm tissue with a Z-interval of 2 μm. Mb, midbrain; Hb, hindbrain. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
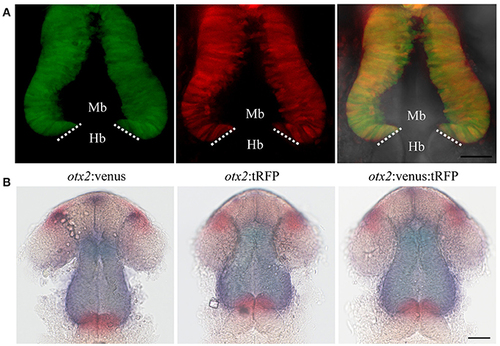
Targeted knock-in of tRFP into the otx2 locus. Expression of tRFP at 24 hpf. Images were taken from live embryos anesthetized in MS-222. (A) Left panel shows a dorsal view with the anterior of the embryo facing upwards, right panel shows a merged image of fluorescent and transmitted light channels. tRFP is expressed in the retina and midbrain, sharply abutting the MHB (dotted line). (B) Lateral view of embryos expressing tRFP. Left panel shows fluorescent channel and right panel shows a merged image of the fluorescent and transmitted light channels. Images are maximum intensity projections covering 50 μm tissue with a Z-interval of 2 μm. MB, midbrain; Hb, hindbrain. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Reporter knock-in into the otx2 locus does not affect endogenous otx2 expression. otx2:venus and otx2:tRFP fish were crossed and sorted into either single (Venus+ or tRFP+) or double positive (Venus+ and tRFP+) embryos at 24 hpf. Images were taken from live embryos anesthetized in MS-222. Panel (A) fluorescent images of the single- and double-positive embryos show no morphological abnormalities. Images are maximum intensity projections covering 50 μm tissue with a Z-interval of 2 μm. Scale bar: 50 μm. Images were taken from live embryos anesthetized in MS-222. Panel (B) Double in situ hybridization for otx2 (blue) and the MHB marker pax2a (red) show no differences in gene expression pattern or morphology of the MHB region between the single- and double-positive groups. Mb, midbrain; Hb, hindbrain. Scale bar 50 μm. EXPRESSION / LABELING:
|
|
Imaging MHB development in real time. To visualize MHB development in real time, embryos from the otx2:tRFP reporter line were mounted and imaged dorsally from 17 to 22 hpf. Tissue sections covering about 40 μm were chosen with a z interval of 2 μm. Images were acquired at 6 min intervals. The otx2:tRFP + cells in the midbrain are clearly visible and cell movements can be followed during ventricular space opening and folding of the neural tube at the MHB. In addition, cellular rearrangements in delaminating neural crest cells and retinal cells are also visible. Time in minutes: seconds. Scale bar: 50 μm. EXPRESSION / LABELING:
|
|
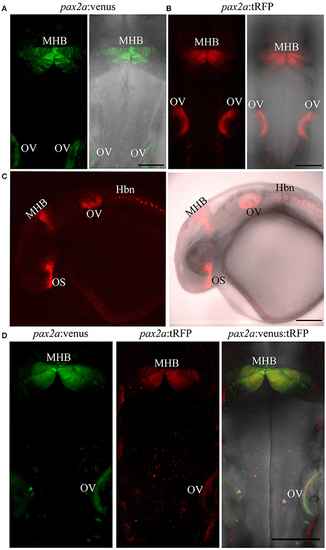
Targeted knock-in of fluorescent reporters into the pax2a locus. Expression of Venus or tRFP driven by the pax2a locus at 24 hpf. Images were taken from live embryos anesthetized in MS-222. (A) Left panel shows dorsal view of Venus expression in the MHB and otic vesicle (OV); right panel shows merged image of fluorescent and transmitted light channels. (B) Left panel shows dorsal view of tRFP expression in the MHB and otic vesicle. (C) Left panel: lateral view of an embryo expressing tRFP in the optic stalk (OS), MHB, OV, and hindbrain neurons (Hbn). Right panel: merged image of fluorescent and transmitted light channels. (D) pax2a:venus and pax2a:tRFP fish were crossed and resulting progeny was sorted into either single (Venus+ or tRFP+) or double positive (Venus+ and tRFP+) embryos at 24 hpf. Compared to single-positive siblings, double positive embryos show no morphological abnormalities. All images are maximum intensity projections covering 50 μm tissue with a Z-interval of 2 μm. Scale bar 100 μm. EXPRESSION / LABELING:
|
|
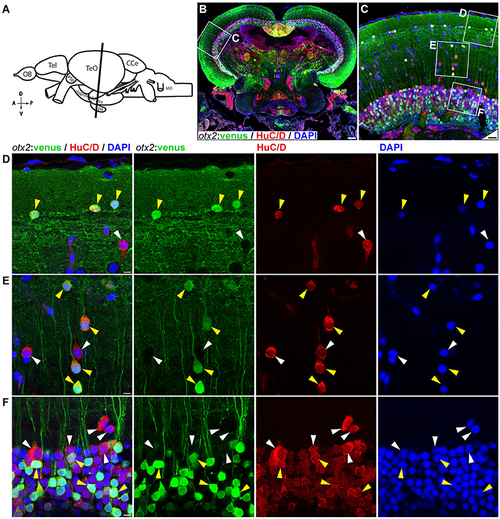
otx2:venus expression in the adult zebrafish midbrain. (A) Schematic representation of the adult zebrafish brain. The position of the cross section of the midbrain with optic tectum, hypothalamus, and pituitary shown in panel (B) is indicated (orientation indicators A->P: anterior to posterior; D->V: dorsal to ventral). (B) Cross section of the midbrain labeled with otx2:venus (green), the pan neuronal marker HuC/D (red) and the nuclear counterstain DAPI (blue). (C) Higher magnification image of the region indicated by white box in panel (B) showing that numerous neurons express otx2:venus. (D–F) Higher magnification images of the regions indicated by white boxes in (C). otx2:venus is co-expressed in HuC/D expressing neurons (yellow arrowheads) in the various tectal sub layers that is characteristic of a teleost midbrain. However, a sub-population of HuC/D positive neurons was negative for otx2:venus (white arrowheads). (D) Type III horizontal neurons that belong to the stratum opticum. (E) Projection neurons that mostly belong to the stratum griseum et album superficiale and centrale regions. (F) Prominent unipolar type XIV interneurons located in the periventricular gray zone of optic tectum. Anatomical descriptions are based on the zebrafish brain atlas (Wullimann et al., 1996), tectal sub layers and neuronal types were interpreted based on Meek (1981) and Meek and Schellart (1978). Scale bars (B): 100 μm; (C): 20 μm; (D–F): 5 μm. (B,C: maximum intensity projection; D–F: single Z-Plane). EXPRESSION / LABELING:
|
|
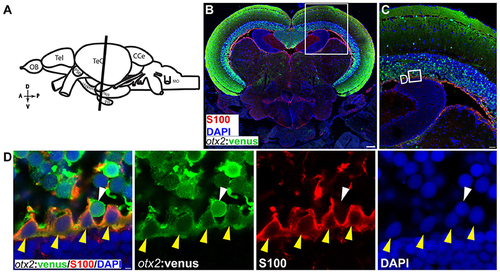
otx2:venus expression in radial glia located at the ventricular zone of the adult zebrafish midbrain. (A) Schematic representation of the adult zebrafish brain. The position of the cross section of the midbrain with optic tectum, hypothalamus, and pituitary shown in panel (B) is indicated (orientation indicators A->P: anterior to posterior; D->V: dorsal to ventral). (B) Cross section of the midbrain labeled with otx2:venus (green), the radial glial marker S100 (red) and the nuclear counterstain DAPI (blue). (C) Higher magnification of the region from midbrain tectum indicated by white boxes in (B). (D) Higher magnification of an inset from panel (C) showing radial glial cells at the ventricular zone of the midbrain tectum labeled by S100 co-expressing otx2:venus. Anatomical descriptions are based on the zebrafish brain atlas (Wullimann et al., 1996). Scale bars (B): 100 μm; (C): 25 μm; (D): 2 (or) 5 μm. (B,C: maximum intensity projection; D: single Z-plane). EXPRESSION / LABELING:
|
|
In situ hybridization showing otx2 expression in the adult zebrafish midbrain. (A,B) In situ hybridization for otx2 was carried out on coronal sections of the adult zebrafish midbrain (scheme shown in A). Otx2 signal resembled reporter expression (Figures 8, 9) in the various tectal layers and particularly strong signals were observed in the periventricular gray zone of the optic tectum. A representative region is the marked with a rectangle and magnified in the adjacent panel (B). Scale bars (A): 100 μm; (B): 20 μm. EXPRESSION / LABELING:
|
|
pax2a:venus expression in the adult zebrafish midbrain. (A) Scheme indicating a plane of coronal section of the adult zebrafish mid brain with optic tectum, hypothalamus and the pituitary corresponding to the cross section in “B” (orientation indicators A->P: anterior to posterior; D->V: Dorsal to Ventral). (B) Cross section of the midbrain immunostained against venus (green) and DAPI (blue). (C) in situ hybridization against pax2a in the midbrain similar to the coronal planes in “A,B” showing similar expression pattern as that of venus in the knock in reporter line. (D,F,H) Higher magnification of a midbrain indicated by white boxes in “B” showing neuronal nuclei expressing venus in regions, valvula cerebelli (red dotted line area) and dorsal tegmental nucleus (yellow dotted line area) “D,” neighboring (left side) to the lateral longitudinal fascicle “F” and the rostral tegmental nucleus “H.” (E,G,H) Higher magnification of a region from the midbrain tectum indicated by black boxes in “C” showing in situ expression pattern for pax2a showing highly similar pattern to their counter parts expressing pax2a:venus. Anatomical descriptions were based on (zebrafish brain atlas; Wullimann et al., 1996). Scale bars (B,C): 100 μm; (D,E,G,I): 20 μm; (F,H): 10 μm. (B: max intensity projection; D,F,H: single Z-Plane; C,E,G,I: DIC image). EXPRESSION / LABELING:
|