- Title
-
Distribution of neurosensory progenitor pools during inner ear morphogenesis unveiled by cell lineage reconstruction
- Authors
- Dyballa, S., Savy, T., Germann, P., Mikula, K., Remesikova, M., Špir, R., Zecca, A., Peyriéras, N., Pujades, C.
- Source
- Full text @ Elife
|
Expansion of the neuroblast delamination domain and formation of the SAG rudiment. (a) Overview of the imaging and image processing strategy: inner ears of zebrafish embryos stained for cell membrane, nucleus and cell fate markers were imaged between 14-42 hpf. Image datasets were processed by nucleus center detection, cell tracking and cell shape segmentation. Data were validated and curated (Figure 1—figure supplement 1). (b–d) Time-lapse stills showing the posterior expansion of the neuroblast delamination domain over time; 3D-rendering of segmented epithelial neuroblasts (green) in context of the otic structure (plasma membranes in magenta) at indicated times; insets display only the segmented delamination domain with the otic vesicle contour in white. ID Dataset: 140210aX; see Figure 1—figure supplement 2d for additional analyses. (e–g) Time-lapse stills showing a segmented delaminating neuroblast (red; Video 2); (e’–g’) magnifications of framed regions in (e–g). ID Dataset: 140426aX. (h–i) Still images from Video 1 displaying: otic tissue architecture (h), and cellular distribution (i) upon SAG formation. Reconstructed cell centers are color-coded according to cell position/identity (see legend). ID Dataset: 140423aX. SAG/ALLg, statoacoustic/anterior lateral line ganglia. AM/PM, anterior/posterior maculae. |
|
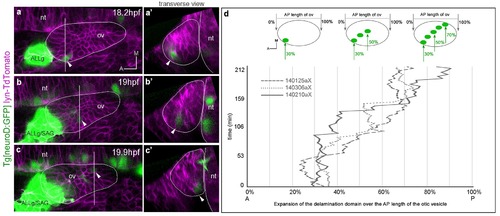
Posterior expansion of the otic neuroblast delamination domain. Tg[neuroD:GFP] embryos were injected with lyn-TdTomato mRNA at 1cell-stage and imaged from 14.5 hpf onwards. Embryos express GFP (green) in neuronal progenitors and differentiating neuroblasts, and TdTomato in all cell membranes (magenta). In the case of the inner ear, GFP is expressed in epithelial neuroblasts just prior to delamination and in the SAG neuroblasts. (a–c) Still image views of the ventral otic vesicle at the indicated time points showing the quick expansion of the delamination domain in the otic epithelium within 2 hr from anterolateral to posteromedial regions; note that at this stage the rudiment of the adjacent ALLg is already visible. (a’–c’) Transverse views are digital reconstructions along the lines indicated in (a–c) and illustrate that the onset of neuroblasts’ delamination progresses from lateral to medial domains (see arrowheads). ALLg, anterior lateral line ganglion; SAG, statoacoustic ganglion; nt, neural tube. The otic vesicle contour is depicted in white. ID Dataset: 140210aX. (d) Plot depicting the posterior expansion of the neuroblast delamination domain as assessed by neuroD-GFP expression. The position of posterior-most neuroD-GFP expressing cells in the otic epithelium, and the anterior and posterior edge of the otic vesicle (dorsal view) were assessed over time (see scheme). The plot displays the position of posterior-most GFP epithelial cells as a percent of the AP otic vesicle length (ID datasets: 140306aX, 140125aX, 140210aX). |
|
The organization of cells within the SAG relies on specific temporal and spatial cues. (a) Flat representation of the neuroblast lineage tree with branches indicating cell divisions. The x-axis displays the time of embryonic development in hours post-fertilization (hpf). Neuroblast lineages are displayed from the moment of delamination onwards and ordered and color-coded according to delamination timing (intervals: 18–20 hpf white, 20–22 hpf yellow, 22–24 hpf orange, 24–30 hpf red). Some cells were not tracked until the end of the sequence, and are depicted as interrupted lines. The extensive cell loss during the early stages of delamination (18–22 hpf) was verified in a second embryo; in both cases, about 25% (23.2% and 26.8%) of the otic epithelial cells at 18 hpf exit by delamination in the consecutive four hours. (c–c',e–e',g–g') Neuroblasts within the SAG (n = 144 of roughly total n = 250) were backtracked to their progenitor state in the epithelium (n = 98; b,d,f; Videos 3 and 5). Cell lineages were color-coded for: time of delamination (b–c'; same intervals as in (a)), position in the epithelium along the AP (d–e'), or ML (f–g') axes. Note that ML organization of neuroblasts within the SAG (c–c') relies on their delamination order, and that the blue/white/red epithelial pattern (d–e'; neuroblasts AP position) but not the green/white/red one (f–g'; neuroblasts ML position) is maintained in the SAG over this time period (18–30 hpf). Reconstructed cell centers were displayed as colored-dots together with the corresponding raw images (plasma membranes in grey level). (b,d,f) dorsal views; (c,e,g) ventral views; (c',e',g') lateral views. Anterior is always to the left. For this analysis, Tg[cldnb:lynGFP] Tg[Isl3:GFP] line was injected with H2B-mCherry mRNA at 1cell-stage (Tables 1–2). ID Dataset: 140426aX; see Figure 2—figure supplement 1 for additional analysis. |
|
Time of delamination and position of epithelial neuroblasts prefigure their location within the SAG. Tg[cldnb:lynGFP] Tg[Brn3c:GFP] embryo was injected with H2B-mCherry at 1cell-stage, imaged and analyzed from 16 hpf (Table 1). Reconstructed cell centers from neuronal progenitors were color-coded for time of delamination (a), or position along the AP axis in the otic epithelium (c), and followed from 18 hpf to 24 hpf. Note that: (i) among the delaminated neuroblasts from 18 hpf to 24 hpf, those delaminating earlier (white cell centers in a-b) are more medially located in the SAG than those delaminating later (purple cell centers in a-b); and (ii) the relative position of neuronal precursors along the AP axis in the otic epithelium is conserved within the SAG (see cyan anterior cells vs. red posterior cells in c-d’). ID dataset: 140423aX. |
|
Spatial distribution of epithelial neuroblasts according to division behavior or delamination time. Tg[cldnb:lynGFP] Tg[Isl3:GFP] embryo was injected with H2B-mCherry at 1cell-stage, imaged and analyzed from 18 hpf to 36.2 hpf (Table 1). Images of (a) nuclei as Maximal Intensity Projection of few orthoplanes of the ventral otic vesicle, and (b) plasma membranes as 3D-rendering are displayed. (c) Spatial distribution of neuroblasts, whose reconstructed cell centers were color-coded according to their division behavior -before or after delamination- (Figure 3). Out of 116 tracked epithelial neuroblasts, 42 divide before delamination (red), 40 do so after delamination (orange), and 34 do not divide within this time window (blue). Note that there is no preferential spatial distribution of cells for these features. (d) Reconstructed cell centers of neuroblasts (n = 131) were color-coded according to time of delamination: 18–20 hpf white, 20–22 hpf yellow, 22–24 hpf orange, 24–30 hpf red. Neuroblasts giving rise to two sister cells falling into distinct delamination intervals are shown as bicolored cell centers (n = 11). ID Dataset: 140426aX. |
|
Spatiotemporal pattern of hair cell differentiation and map of sensory progenitors. Differentiated hair cells were tracked during 18 hr in control and MO-neurog1, and reconstructed cell centers were color-coded according to the differentiation time displayed in the legend (Video 6). (a–b,f–g) Spatiotemporal pattern of hair cell differentiation of the anterior/posterior maculae (AM/PM); reconstructed colored cell centers overlaid with the corresponding raw images (hair cell fate in grey level) from Tg[Brn3c:GFP] embryos; (c,h) reconstructed colored cell centers in lateral view. Note how the temporal but not the spatial development is altered in the MO-neurog1 PM (see Figure 4—figure supplement 1). (d,i) Map of hair cell progenitors in the whole otic vesicle (Videos 7-8); the maps were generated by backtracking the differentiated PM hair cells (e,j). ID Datasets: 140507aX for control, 140519aX for MO-neurog1; see Figure 4—figure supplement 1 for additional analyses |
|
Temporal pattern of hair cell differentiation in AM and PM. Tg[Brn3c:GFP] embryos injected with H2B-mCherry mRNA at 1cell-stage (with/without MO-neurog1) were imaged from 24 hpf to 42 hpf. (a) Graphs showing the increase of differentiated hair cells in the anterior (AM) and posterior (PM) maculae over time; the final number of differentiated hair cells at 42 hpf is indicated. Each line corresponds to a differentiated hair cell plotted from and color-coded for time of differentiation (see legend). Tether cells are depicted as white lines. Note the temporal differences in the development of the PM between control and MO-neurog1 embryos, with no major changes in the spatial pattern (Figure 4). Asterisk depicts an apoptotic hair cell. ID Datasets: 140507aX for control, 140519aX for MO-neurog1. (b) Spatiotemporal pattern of hair cell differentiation of the AM and PM of both ears of Tg[Brn3c:GFP]Tg[Isl3:GFP] embryo injected with lyn-TdTomato and H2B-mCherry mRNA at 1cell-stage (imaged from 25 hpf to 36 hpf). Differentiated hair cells were tracked and reconstructed colored cell centers overlaid with the corresponding raw images (hair cell fate/SAG in grey level as 3D-rendering). Graphs showing the increase of differentiated hair cells in the AM and PM over time; the final number of differentiated hair cells at 36 hpf is indicated. ID Dataset: 140402aX. (c) Spatiotemporal pattern of hair cell differentiation of AM and PM. Tg[Brn3c:GFP] embryo injected with lyn-TdTomato mRNA at 1cell-stage and imaged from 25 hpf to 45 hpf. Graphs displaying the increase of differentiated hair cells in AM/PM. Hair cell progenitors and differentiated hair cells were tracked (except for the one encircled in blue), and reconstructed colored cell centers overlaid with the corresponding raw images (MIP of lynTomato and GFP signal of a few slices in grey level). ID Dataset: 130326aX. Legend in panel (a) applies to all plots and diagrams of the figure. |
|
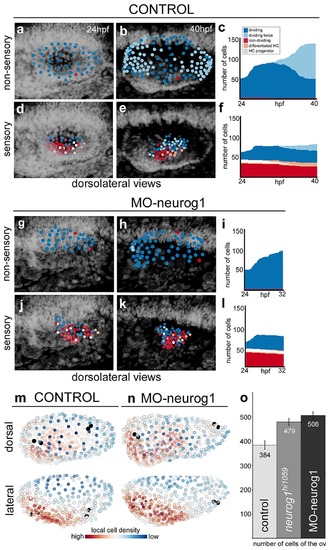
Heterogeneous cell behavior in the non-sensory and sensory domains. Neighboring cells in the non-sensory and sensory domains of control (a–f) and MO-neurog1 (g–l) were tracked and reconstructed cell centers were color-coded according to cell proliferation/differentiation status (see legend in (c); Video 9); they were plotted on the top of the corresponding raw images (a–b,d–e,g–h,j–k; nuclei in grey level), or in graphs over time (c,f,i,l) displaying the total number of cells in each domain and their status in the course of the video. Note the differences in the graphs between non-sensory and sensory domains, but not between control and MO-neurog1 embryos. (m–n) Estimated local cell densities at 24 hpf are represented by color-coded cell centers across the whole otic epithelium (Video 10). Tg[Brn3c:GFP] embryos injected with H2B-mCherry and with/without MO-neurog1 at 1cell-stage were used for full lineage reconstruction (Tables 1–2). Anterior is always to the left. ID Datasets: 140507aX for control, 140519aX for MO-neurog1; see Figure 5—figure supplement 1 for additional analyses. (o) Graphic depicting the total number of cells in the otic vesicles for wild type (control, n = 3), neurog1hi1059/ hi1059 mutant in the Tg[Isl3:GFP] background (n = 3), and MO-neurog1 embryos (n = 2) at 24 hpf. |