- Title
-
TAEL: A zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control
- Authors
- Reade, A., Motta-Mena, L.B., Gardner, K.H., Stainier, D.Y., Weiner, O.D., Woo, S.
- Source
- Full text @ Development
|
Optimization of TAEL as an optogenetic gene expression system for zebrafish. (A) Schematic depicting general mechanism for single-protein component, LOV-based, light-responsive gene expression systems. (B) Schematic depicting transcriptional activators. (C,D) Percentage of unaffected, healthy embryos per amount of mRNA injected. Injection of GAVPO or EL222 mRNA is significantly more toxic compared with the equivalent amount of GFP control mRNA (C), but GAVTA and TAEL mRNA injections produce less toxicity (D). *P<0.05, **P<0.0001, two-tailed Fisher's exact test. GFP data in D reproduced from C. Data represent mean percentage (±s.e.m.) of total embryos (n) from 2-3 independent experiments. GAVPO, n=849 embryos; EL222, n=1421 embryos; GFP, n=1190 embryos; GAVTA, n=809 embryos; TAEL, n=824 embryos. (E-G) TAEL induces robust mCherry transcription from the C120 promoter at 24 hpf (G,G′), with no visible induction of mCherry expression in dark (F,F′) or uninjected controls (E,E′). Scale bars: 200 µm. (H) qPCR analysis of TAEL-induced mCherry expression over time, normalized to 0 h post-activation for each illumination pattern. Pulsed illumination (purple line) results in stronger induction compared with constant illumination (green line). Red dotted line shows that mCherry transcription quickly turns off after activated embryos are returned to dark conditions. Embryos kept in the dark for all time points exhibit baseline mCherry expression (orange line). Data represent means±s.e.m. from three biological replicates, each with three technical replicates. Y-axis is displayed on a log2 scale. |
|
Spatial control of TAEL induction achieved using four different imaging modalities. Left column shows activation region (blue) for each method. Middle and right columns show resulting mCherry expression (red) 4 h post-activation. (A,C,E,G,I,K) Bright-field and 488 nm channels merged. (B,D,F,H,J,L) Bright-field and 561 nm channels merged. (B′,D′,F′,H′,J′,L′) 561 nm channel only. Unless otherwise noted, images are lateral views with rostral to the left. (A-B′) Closing down the field diaphragm on an epifluorescence microscope (488 nm, GFP excitation setting) restricts the light coming through the objective and illuminates the sample with a small hexagonal column. (C-D′) Region of interest (ROI) on a point scanning confocal to restrict scanning of the 488 nm laser to a small square. (E-H′) Digital micromirror device (DMD) illuminated with a 470 nm LED to project variously sized square columns of blue light onto the embryo. (I-L′) Restricted scanning range of the 488 nm laser on a digital scanned laser light sheet microscope (DSLM) to project variously wide beams of blue light through the embryo. (I) En face view with head and tail as indicated. (J-J′) Dorsal view with anterior to the bottom. Brackets (L,L′) indicate an example of ‘off-target’ mCherry expression. Arrows in I and K indicate position of the light sheet. Scale bars: 100 µm. |
|
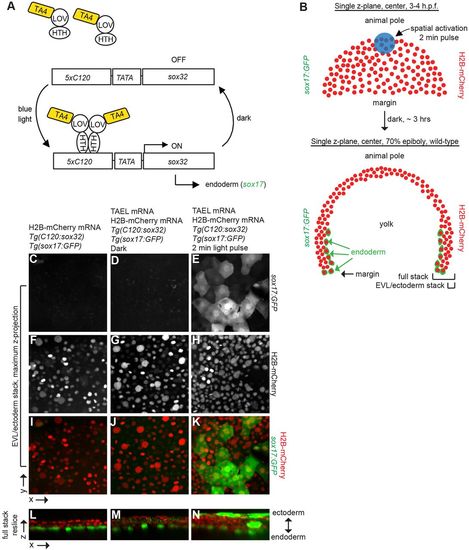
Conversion of presumptive EVL and ectoderm to endoderm via TAEL-dependent Sox32 expression. (A) Schematic depicting experimental set-up for light-induced endoderm induction. Tg(cryaa:Venus;C120:sox32); Tg(sox17:GFP) embryos were injected with TAEL mRNA. Ectoderm progenitors at the animal pole were illuminated with blue light, leading to misexpression of sox32 and adoption of endoderm fate (expression of sox17:GFP). (B) Top panel depicts an embryo at 3-4 hpf with nuclei labeled by H2B-mCherry (red). Activating light was restricted to the animal pole (blue circle, approximately 116 μm in diameter). Bottom panel depicts a transverse cross section of an embryo at 70-80% epiboly. Endodermal cells are labeled by Tg(sox17:GFP) (green) and all nuclei are labeled with H2B-mCherry (red). Brackets labeled ‘EVL/ectoderm stack’ and ‘Full stack’ indicate approximate positions of images shown in C-K and L-N, respectively. (C-K) Maximum z-projections of EVL and ectoderm layers only. Embryos lacking TAEL (C) or expressing TAEL but kept in the dark (D) show no GFP-positive cells in these layers. However, activated embryos (E) exhibit numerous GFP-positive cells within the EVL and ectoderm domain. (L-N) Re-sliced x-z views of the entire acquired stack (EVL/ectoderm towards the top, endoderm toward the bottom). In embryos lacking TAEL (L) or expressing TAEL but kept in the dark (M) GFP-positive cells are restricted to a single endodermal layer adjacent to the yolk, whereas in activated embryos (N), GFP-positive cells are located in multiple cell layers, especially in the outer EVL and ectoderm domains. Uninjected, 17% with ectopic endoderm, n=17; dark, 33% with ectopic endoderm, n=9; light activated, 65% with ectopic endoderm, n=17. |
|
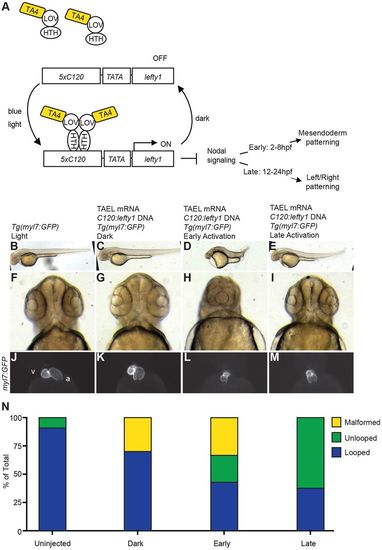
Temporal control of Nodal signaling via TAEL-induced expression of lefty1. (A) Schematic depicting experimental set-up for TAEL-induced Nodal inhibition. Embryos were injected with a transgene containing the C120 promoter driving expression of the Nodal antagonist lefty1 along with TAEL mRNA. Embryos were then globally illuminated with blue light either from 2-8 hpf (early) or 12-24 hpf (late). (B-E) Lateral views of embryos at 48 hpf show that early activation results in severely shortened embryonic axis (D) whereas late activation does not affect body length (E). (F-I) Rostral views of 48 hpf embryos show that early (H) but not late (I) activation produces cyclopia, indicative of loss of cephalic mesoderm. (J-M) Ventral views of 48 hpf embryos with hearts labeled by Tg(myl7:GFP) expression. Control embryos exhibit asymmetric heart looping (J,K). In contrast, late activation of lefty1 expression produces unlooped hearts with both chambers located at the midline (M). Early activation produced both unlooped and malformed hearts (L). A, atrium; V, ventricle. (N) Quantification of heart defects. Uninjected, n=55; dark, n=10; early, n=21; late, n=24. |
|
Light-induced gene editing in zebrafish. Light-induced gene knockout can be achieved by combining the TAEL system with the CRISPR/Cas9 system. (A) Three promoter construct containing: (1) the heart-specific promoter myl7 in front of GFP to label injected embryos, (2) Cas9 under the control of the C120 promoter and (3) a U6 promoter in front of the guide RNA sequence targeting tyrosinase, a protein essential for pigment formation. TAEL mRNA was co-injected with the construct depicted in A into wild-type single-celled embryos. Embryos were illuminated globally with 465 nm blue light and then scored for disruption in pigment formation at 2 dpf. (B-D) Wild-type, uninjected embryos (B) exhibit normal pigmentation as do injected dark controls (C). Pigment formation is disrupted in injected and illuminated embryos (D, arrow). Approximately 90% of illuminated embryos (n=18) showed various levels of pigment disruption, while 100% of dark control embryos examined (n=11) exhibited pigmentation comparable to wild-type uninjected embryos. (E) Representative CEL I nuclease assay indicates mutagenesis of the tyr locus in illuminated but not dark or uninjected control embryos. Arrows indicate cleavage fragments. Scale bar: 100 µm. |
|
LightOn system induces kaede expres-sion from the UAS promoter in a light-gated manner. A-C. Tg(UAS:kaede) embryos were injected with GAVPO mRNA and then illuminated with con-stant, global blue light (465 nm) from approximately 4-24 hpf (hours post-fertilization). GAVPO injected embryos showed robust induction of Kaede fluores-cent protein at 24 hpf (C), with no visible induction of Kaede expression in dark (B) or uninjected controls (A). D-F. Following blue light illumination, GAVTA is unable to induce transcription of kaede from the UAS promoter (F, F'). |