- Title
-
Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish
- Authors
- Xiao, C., Gao, L., Hou, Y., Xu, C., Chang, N., Wang, F., Hu, K., He, A., Luo, Y., Wang, J., Peng, J., Tang, F., Zhu, X., Xiong, J.W.
- Source
- Full text @ Nat. Commun.
|
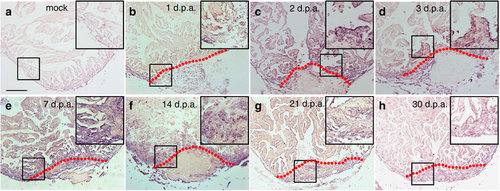
brg1 is upregulated during cardiac regeneration in zebrafish. In situ hybridization was performed on paraffin sections of mock-operated zebrafish (a) and those with amputated ventricular apexes (b–h) at the indicated time points using a digoxigenin-labelled anti-sense brg1 RNA probe. Note induced expression of brg1 in the injured heart from 1 to 14 d.p.a. (b–f). Dashed lines mark the resection sites; the right upper corner is high-magnification image of the framed area; similar results were confirmed by performing three independent experiments. Scale bar, 100 μm. |
|
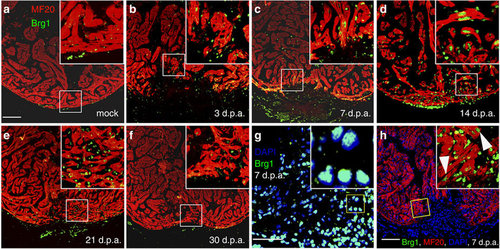
Brg1 is activated in multiple types of cells during cardiac regeneration in zebrafish. (a–f) Immunofluorescence staining of Brg1 and cardiac sarcomere myosin heavy chain (MF20) was performed on paraffin sections of mock-operated zebrafish (a) and those with amputated ventricular apexes (b–f) at the indicated time points. The right upper corners are high-magnification images of the frame area in a–f, showing Brg1 co-localization in MF20-positive myocytes. (g) Co-staining of Brg1 and 4,6-diamidino-2-phenylindole (DAPI) in paraffin sections of amputated apexes at 7 d.p.a. The right upper corner is high-magnification image of the framed area. (h) Immunofluorescence staining of Brg1 and MF20 of amputated heart at 7 d.p.a., showing the co-localization of Brg1 and MF20. The right upper corner is high-magnification image of the framed area. These data were confirmed by performing three independent experiments. Scale bars, 100 μm. |
|
Inhibition of brg1 impairs cardiac regeneration. (a–d) Representative sections from wild-type siblings (a,c) and Tg(hsp70:dn-xbrg1) (b,d) hearts at 30 d.p.a., evaluated by AFOG staining (a,b), and immunofluorescence staining with anti-myosin heavy chain (MF20) (c,d). Note massive fibrosis (b) and compromised myocardial regeneration (d) in Tg(hsp70:dn-xBrg1) hearts (tg). Dashed lines mark the resection site. (e–g) Paraffin sections of 14 d.p.a. regenerating heart of wild-type sibling (e) and Tg(hsp70:dn-xBrg1) (f) hearts co-stained for BrdU (green), Mef2C (red) and 4,6-diamidino-2-phenylindole (DAPI; blue). Higher-magnification images of areas in squares are shown in the upper-right corners, and Mef2C+/BrdU+ double-positive cardiomyocytes are indicated by arrowheads. (g) Percentages of Mef2C+/BrdU+ cardiomyocytes in the injured area (***P<0.001; n=6 for siblings and 7 for transgenic hearts; data are mean percentages±s.e.m., paired Student’s t-test). (h–j) Paraffin sections of 14 d.p.a. wild-type Tg(gata4:EGFP) sibling (h) and Tg(hsp70:dn-xbrg1; gata4:EGFP) (i) hearts stained with anti-EGFP and DAPI. The average of fluorescence intensity was calculated using Imaris software (j) (**P<0.01; n=6; data are mean percentages±s.e.m.; paired Student’s t-test). Scale bars, 100 μm. |
|
cdkn1a and cdkn1c are induced and enriched in the myocardium of Tg(hsp70:dn-xBrg1) transgenic hearts. (a–h) RNAscope in situ hybridization analysis with cdkn1a (a–d) and cdkn1c (e–h) probes on frozen sections of uninjured wild-type (WT) hearts (a,e), injured WT hearts at 3 d.p.a. (b,f), injured WT sibling hearts at 14 d.p.a. (c,g) and injured Tg(hsp70:dn-xBrg1) transgenic hearts at 14 d.p.a. (d,h). Note the robust induction of cdkn1a (d) and cdkn1c (h) induction in Tg(hsp70:dn-xBrg1) transgenic hearts compared with WT sibling hearts at 14 d.p.a. after heat shock. Black arrowheads indicate the cdkn1a or cdkn1c signals. The panels below a–h are higher-magnification images of areas in squares of a–h. (i–p) Bright-field images of cdkn1a (i–l) and cdkn1c (m–p) expression by RNAscope merged with immunostaining signal images of MF20 on frozen sections of uninjured WT hearts (i,m), injured WT hearts at 3 d.p.a. (j,n), injured WT sibling hearts at 14 d.p.a. (k,o) and injured Tg(hsp70:dn-xbrg1) transgenic hearts at 14 d.p.a. (i,p). Higher-magnification images of squared areas of i–p are shown below their respective panels. Note that both cdkn1a (i) and cdkn1c (m) are normally expressed in cardiomyocytes in uninjured WT hearts, and that they are highly induced in MF20-positive cardiomyocytes of Tg(hsp70:dn-xBrg1) transgenic hearts (l,p) compared with WT sibling hearts (k,o) at 14 d.p.a. White arrowheads show cdkn1a or cdkn1c signals in cardiomyocytes. Tg, Tg(hsp70:dn-xBrg1); (+) HS, heat shock; Scale bars, 100 μm. |
|
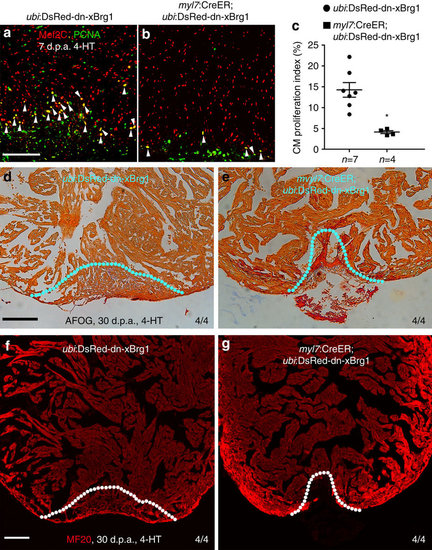
Myocardial-specific inhibition of Brg1 interferes heart regeneration. (a–c) PCNA+/Mef2C+ proliferating cardiomyocytes decreased in Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts (b) compared with control Tg(ubi:DsRed-dn-xBrg1) transgenic hearts (a) at 7 d.p.a. Statistics of cardiomyocyte proliferation index is shown (*P<0.05; data presented are mean±s.e.m.; paired Student’s t-test) (c). White arrowheads point to PCNA+/Mef2C+ proliferating cardiomyocytes; n, the number of hearts analysed; ubi:DsRed-dn-xBrg1 stands for Tg(ubi:loxP-DsRed-STOP-loxP-dn-xBrg1); tamoxifen (4-HT) was applied at 3 days before injury. (d,e) AFOG staining revealed accumulated fibrin and fibrosis in Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts (e) compared with control Tg(ubi:DsRed-dn-xBrg1) transgenic hearts (d) at 30 d.p.a. (f,g) MF20 staining showed compromised myocardial regeneration in Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts (g) compared with control Tg(ubi:DsRed-dn-xBrg1) transgenic hearts (f) at 30 d.p.a. 4/4, all 4 hearts analysed showed the same phenotype. Scale bars, 100 μm. |
|
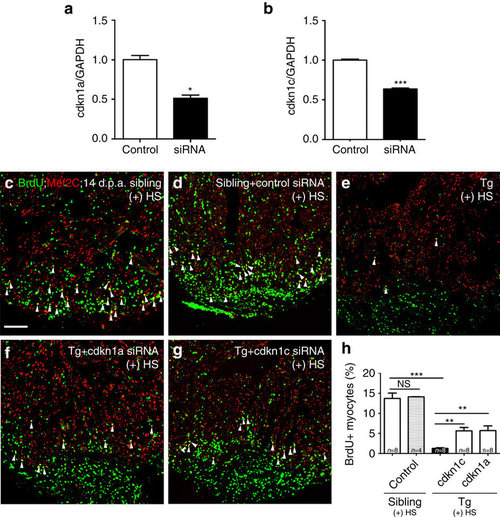
siRNA knockdown of either cdkn1a or cdkn1c partially rescues proliferating cardiomyocytes in the Tg(hsp70: dn-xBrg1) heart. (a,b) Quantitative PCR showed that nanoparticle-encapsulated siRNA efficiently decreased the RNA levels of cdkn1a and cdkn1c in wild-type hearts at 2 d.p.a., into which control and cdkn1a (a) or cdkn1c (b) siRNA were injected at 1 d.p.a. The RNA level was normalized to GAPDH (*P<0.05, ***P<0.001; data presented are mean±s.e.m.; paired Student’s t-test). (c–h) Ventricular apex amputation was performed in wild-type siblings and Tg(hsp70:dn-xBrg1) zebrafish, followed by heat shock treatment for 30 min daily from 5 to 14 d.p.a. The Mef2C+/BrdU+ double-positive cardiomyocytes were comparable in control siRNA-injected (d) and uninjected (c) hearts. Either encapsulated cdkn1a (f) or cdkn1c (g) siRNA partially rescued the ratio of Mef2C+/BrdU+ double-positive cardiomyocytes in Tg(hsp70:dn-xBrg1) hearts compared with those in uninjected control transgenic hearts (e). Scale bar, 100 μm. (h) Statistics of c–g (**P<0.01, ***P<0.001; data are mean±s.e.m.; one-way analysis of variance followed by Bonferroni’s multiple comparison test). The number (n) of hearts analysed in each group is indicated in each bar. |
|
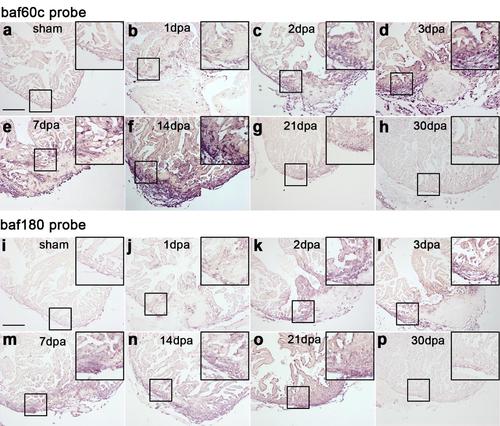
Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and those with amputated ventricular apices from 1 to 30 dpa (b-h and j-p) with digoxigenin-labeled baf60c probe (a-h) or baf180 probe (i-p). Higher-magnification images of areas in squares were shown in the upper-right corners of panels a-p. Scale bars, 100 μm. Representative data from 3 independent experiments (n=5 hearts). |
|
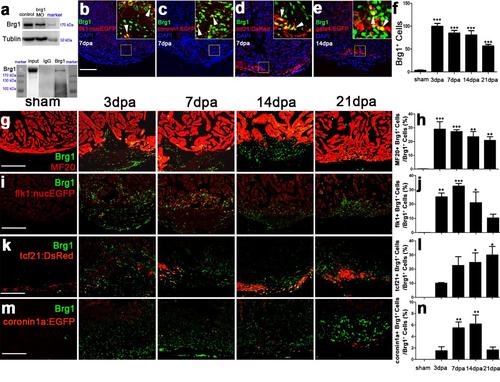
Brg1 is activated in multiple types of cells during cardiac regeneration. (a) Upper panel, western blot with anti-Brg1 antibody showing that Brg1 decreased in brg1 morphant embryos compared with control embryos at 48 hpf. Tubulin served as a loading control. Lower panel, Immunoprecipitation (IP) by anti-Brg1 antibody showing that Brg1 antibody was able to pull down the endogenous Brg1 of adult zebrafish hearts at 7 dpa. (b-e) Immunostaining of Brg1 and EGFP or DsRed on paraffin sections of injured hearts, in which the endocardium/endothelium were labeled by Tg(flk1:nucEGFP) at 7 dpa (b), macrophages and neutrophils by Tg(coronin1a:EGFP) at 7 dpa (c), the epicardium by Tg(tcf21:DsRes) at 7dpa (d), and the myocardium by Tg(gata4:EGFP) at 14 dpa (e). In the upper-right corners of panels b-e, higher magnification images show that Brg1 was located in the endocardium (b), macrophages/neutrophils (c), the epicardium (d), and the myocardium (e). (f) Quantification of Brg1-positive cells of sham and injured hearts from 3 to 21 dpa. (g-n) Immunostaining of Brg1 and MF20 (g), Brg1 and flk1:nucEGFP (i), Brg1 and tcf21:DsRed (k), as well as Brg1 and coronin1a:EGFP (m) of sham and injured hearts from 3 to 21 dpa. Quantification of Brg1+ cells co-expressing MF20 (h), Brg1+ cells co-expressing flk1:nucEGFP (j), Brg1+ cells co-expressing tcf21:DsRed (i), and Brg1+ cells co-expressing coronin1a:EGFP (n). Scale bars,100 μm. For all quantifications, data are mean ± s.e.m.; one-way ANOVA followed by Dunnett’s Multiple Comparison Test, *p <0.05, **p <0.01, ***p <0.001 |
|
Overexpression of Xenopus dominant-negative Brg1 (dn-xBrg1) inhibits brg1 function in zebrafish. (a) Schematic of the Tol2-based construct of conditional expression of dn-xBrg1 driven by the heat-shock promoter 70 (hsp70). (b) Western blots showing that heat-induced dn-xBrg1 protein increased in transgenic adult hearts (tg) compared with those in non-transgenic wild-type siblings (wt) with (+HS) or without heat shock (-HS), or transgenic adult hearts without heat shock (-HS). Alpha-actin served as a loading control. (-) HS, without heat shock; (+) HS, with heat shock. (c-f) Heart tube became stenotic and abnormal looping in tg embryos compared with wt sibling embryos at 48 hpf after heat shock (30 min each time at 5 hpf, 17 hpf, 29 hpf, and 41 hpf) or wt and tg embryos without heat shock. The heart tube was labeled by in situ hybridization with cmlc2, vmhc, amhc, or nppa probes. In situ hybridization showing that cardiac genes bmp4, tbx2b, and notch1b were abnormally expressed in tg embryos compared with wt sibling embryos after heat shock (30 min each time at 9 hpf, 33 hpf and 57 hpf) (g, h, i). Note the expanded bmp4 (g) and tbx2b (h) domains and decreased expression of notch1b in the atrioventricular canal of tg embryos (i). (j) Heart tube, labeled by cmlc2, became stenotic but the atrium and ventricle were specified in tg embryos compared with wt sibling embryos at 60 hpf after heat shock. Red brackets indicate expression domains of atrioventricular canal markers (bmp4 and tbx2b); black arrowheads point to decreased expression of notch1b in tg embryos; number of the right-upper corners showing the number of phenotypic embryos out of the total embryos analyzed. Scale bars, 100 μm. |
|
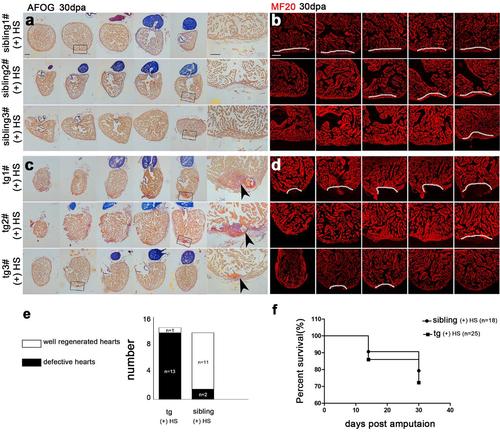
Inhibition of brg1 impairs cardiac regeneration. (a-d) Three wild-type (wt) sibling hearts (a-b) and 3 dn-xBrg1 transgenic (tg) hearts (c-d) at 30 dpa with heat shock treatment from 5dpa to 30dpa were subjected to serial sections, and half of sections were then used for AFOG staining (a, c) and half for MF20 immunofluorescence staining (b, d). Higher-magnification images of areas in squares were shown in the right side of panels a and c. Note cardiac fibrosis (black arrowheads) and compromised myocardial regeneration (dashed lines) in tg hearts (c, d) compared with perfect heart regeneration in wt sibling hearts (a, b). Scale bars, 100 μm. (e) Quantification of defective hearts; the number of well-regenerated hearts (white) or defective hearts (black) in each group was indicated in each bar (n=13 for sibling and n=14 for dn-xBrg1 transgenic total hearts). (f) Heat-induced lethality of wt siblings (n=18) and tg zebrafish (n=25) after heat shock at 14 dpa and 30 dpa. |
|
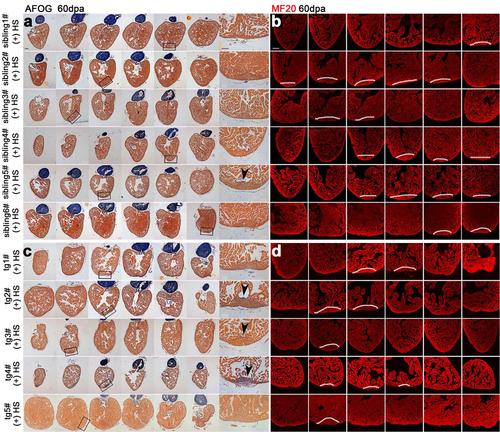
Inhibition of Brg1 causes permanent defects in heart regeneration. Six wild-type (wt) sibling hearts (a-b) and 5 dn-xBrg1 transgenic (tg) hearts (c-d) at 60 dpa (heat shock treatment only from 5dpa to 30dpa) were subjected to serial sections, and half of sections were then used for AFOG staining (a, c) and half for MF20 immunofluorescence staining (b, d). Higher-magnification images of areas in squares were shown in the right side of panels a and c. Note that 3 of 5 tg hearts had fibrosis and compromised myocardial regeneration even although they had no heat-induced dn-xBrg1 proteins from 30 to 60 dpa. Arrowheads indicate fibrosis. Scale bars, 100 μm. |
|
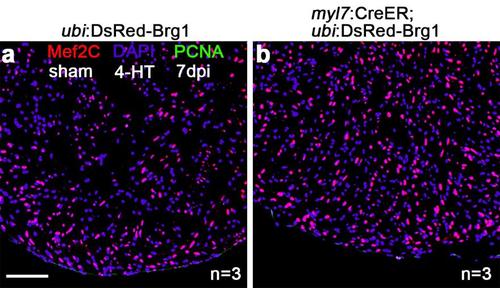
Cardiomyocyte-specific overexpression of brg1 is not sufficient to induce myocyte proliferation. PCNA+/Mef2C+ proliferating cardiomyocytes were not increased in uninjured Tg(myl7:CreER; ubi:loxP-DsRed-STOP-loxP-Brg1) transgenic hearts (b) compared with uninjured control Tg(ubi:loxP-DsRed-STOP-loxP-Brg1) hearts (a). Hearts were induced by tamoxifen for 24hr, and 7 days post inducement paraffin heart sections were costained for PCNA (green), Mef2C (red) and DAPI (purple). n=3; Scale bar, 100μm. |
|
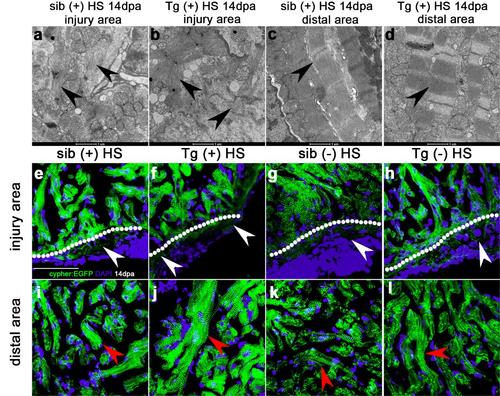
Inhibition of Brg1 has no effect on cardiac sarcomere disassembly during heart regeneration. (a-d) Transmission electron microscopy images of myocytes of wt sibling (sib) (a, c), and Tg(hsp70:dn-xBrg1) transgenic (tg) heart (b, d) at 14 dpa. Note the normal sarcomere structures (black arrowhead) in distal cardiomyocytes from the injury site (c, d) and sarcomere disarray (black arrowhead) in cardiomyocytes in the injury site of both tg and wt hearts with heat shock treatment (a, b). Scale bars, 1μm. (e-h) Z-disks were labeled by using cypher-EGFP fusion protein. Cypher-EGFP-labeled sarcomere was all disarrayed in cardiomyocytes near the injury site (e-h), but was normal in distal area (i-l), of tg and wt sibling hearts at 14 dpa with or without heat-shock treatment. White arrowheads indicate sarcomere disarray (e-h) while red arrowheads indicate normal sarcomeres (i-l). DAPI co-stained for the nuclei; Scale bar, 50μm. |
|
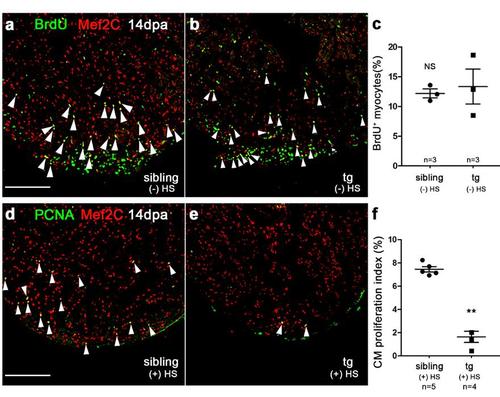
Inhibition of brg1 impairs cardiac regeneration. (a-c) BrdU+/Mef2C+ proliferating cardiomyocyte were comparable between wt sibling hearts (a) and dn-xBrg1 transgenic hearts (b) at 14 dpa without heat shock treatment [(-) HS]. (c) Percentages of BrdU+/Mef2C+ cardiomyocytes in the injured area. (d-f) PCNA+/ Mef2C+ proliferating cardiomyocytes (arrowheads) decreased in dn-xBrg1 tg hearts (e) compared with wt sibling hearts (d) at 14 dpa with heat shock treatment. (f) Quantification of cardiomyocyte proliferation assessed by PCNA+/ Mef2C+ staining Scale bars, 100 μm. Data presented are mean ± s.e.m.; paired Student’s t-test, sample numbers are listed under each group, ** p < 0.01. |
|
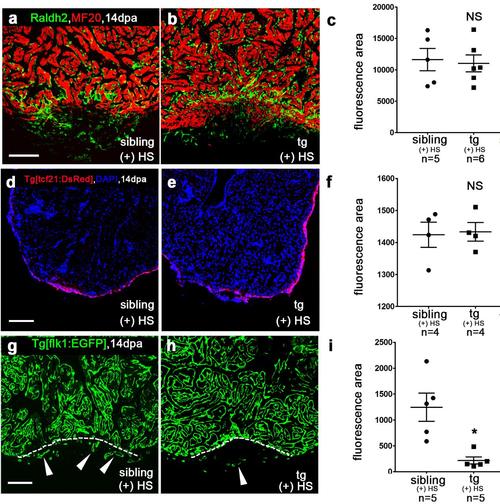
(a-c) Levels of Raldh2 assessed by immunostaining with anti-Raldh2 (green) and anti-myosin heavy-chain (MF20) (red) were comparable in wild-type sibling (a) and Tg(hsp70:dn-xBrg1) (b) hearts at 14 dpa. (c) Quantification of the fluorescence intensity of Raldh2 signals in the original wound sites. (d-f) tcf21:DsRed signal was comparable in Tg(tcf21:DsRed) control hearts (d) and Tg(hsp70:dn-xBrg1;tcf21:DsRed) transgenic hearts (e) at 14dpa. (f) Quantification of the fluorescence intensity of tcf21-DsRed signals was shown (g-i) Coronary vessels were reduced in Tg(hsp70:dn-xBrg1;flk1:EGFP) transgenic hearts (h) compared with those in wild-type Tg(flk1:EGFP) sibling hearts (g) at 14 dpa. The resection sites are marked with dashed lines. (i) Quantification of the fluorescence intensity of flk1-EGFP signals in the original wound sites was shown Heat shock was applied from 5 to 14 dpa. Scale bars, 100μm. Data presented are mean ± s.e.m., paired Student’s t-test); NS, no significant difference; *p <0.05, sample numbers are listed under each group. |
|
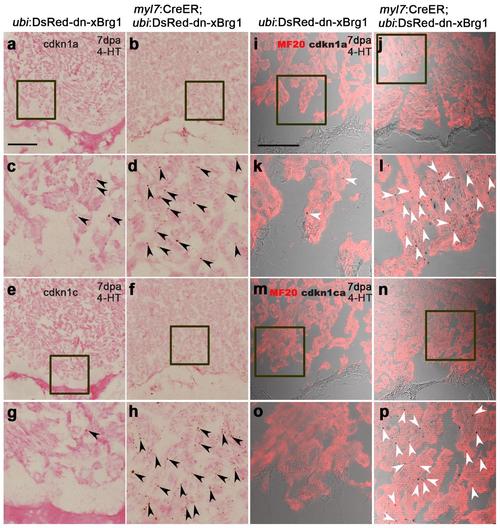
cdkn1a and cdkn1c are induced in Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts. (a-h) RNAScope in situ hybridization analysis with cdkn1a (a-d) and cdkn1c (e-h) probes on frozen sections of injured control Tg(ubi:DsRed-dn-xBrg1) hearts (a, c, e, g) and injured Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts (b, d, f, h) at 7dpa after 4-HT induction. Panels c, d, g and h are high-magnification images of areas in squares in panels a, b, e and f. Black arrowheads indicate the RNAScope signals. (i-p) Bright-field views of cdkn1a (i-l) and cdkn1ca (m-p) expression by RNAscope in situ hybridization, which were merged with MF20 antibody confocal images, on frozen sections of injured Tg(ubi:DsRed-dn-xBrg1) hearts (i, k, m, o) and injured Tg(myl7:CreER; ubi:DsRed-dn-xBrg1) transgenic hearts (j, l, n, p) at 7dpa after 4-HT induction. Panels k, l, o and p are high-magnification images of areas in squares in panels i, j, m and n. White arrowheads show RNAScope signals in cardiomyocytes. Scale bars, 100 μm. |
|
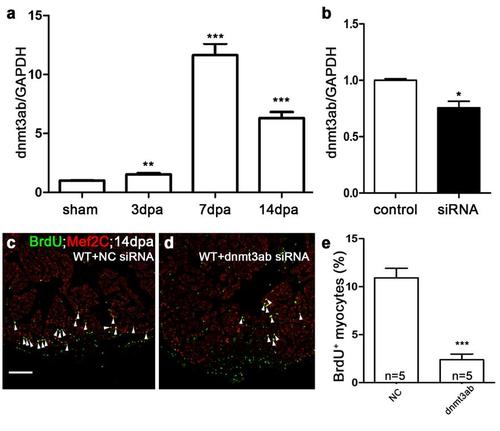
Dnmt3ab is required for myocardial proliferation. (a) Quantitative PCR showed that dnmt3ab was induced in injured hearts at 3, 7, and 14 dpa compared with sham hearts (**P <0.01; ***P <0.001; data presented are mean ± s.e.m.; paired Student’s t-test). (b) Quantitative PCR showed that nanoparticle-mediated dnmt3ab siRNA decreased the RNA level of dnmt3ab in wild-type hearts (*P <0.05; data presented are mean ± s.e.m.; unpaired Student’s t-test). (c-e) BrdU +/Mef2C+ proliferating cardiomyocytes decreased in dnmt3ab siRNA hearts (d) compared with negative control (NC) siRNA heart (c) at 14 dpa. The number (n) of hearts analyzed in each group is indicated in each bar (***P <0.001; data presented are mean ± s.e.m.; paired Student’s t-test). Scale bar, 100μm. |
|
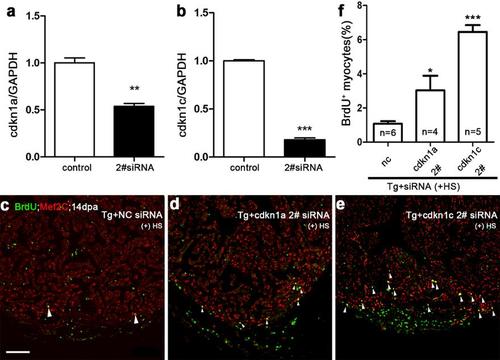
Simultaneous siRNA knockdown of either cdkn1a or cdkn1c increases myocardial proliferation in dn-xBrg1 transgenic hearts. (a, b) Quantitative PCR showed that another independent siRNA for cdkn1a (a) or cdkn1c (b) decreased the RNA levels of cdkn1a and cdkn1c in wild-type hearts at 2 dpa. Control, cdkn1a 2# (a), or cdkn1c 2# (b) siRNA were injected at 1 dpa. The RNA level was normalized to GAPDH (**p <0.01, ***p < 0.001; data presented are mean ±s.e.m.; paired Student’s t-test). (c-f) BrdU+/Mef2C+ proliferating cardiomyocytes increased in dn-xBrg1 transgenic hearts at 14 dpa injected with either cdkn1a 2# (d) or cdkn1c 2# (e) compared with control NC siRNA (c). Statistics of panels c-e is shown (f) (*p <0.05, ***p <0.001; data are mean ± s.e.m.; one-way ANOVA followed by Dunnett’s Multiple Comparison Test; nc served as control). The number (n) of hearts analyzed in each group is indicated in each bar; heat shock was applied from 5 to 14 dpa. Scale bar, 100μm. |
|
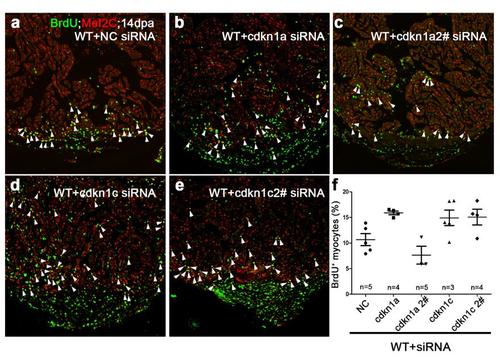
Knockdown of CDK inhibitors in wild-type hearts had minimal effects on myocardial proliferation. BrdU+/Mef2C+ proliferating cardiomyocytes (white arrowheads) were comparable among negative control (NC) siRNA (a), cdkn1a siRNA (b), cdkn1a 2# siRNA (c), cdkn1c siRNA (d), or cdkn1c 2# siRNA-treated (e) wild-type hearts at 14 dpa, consistent with low-level of cdkn1a and cdkn1c in wild-type injured hearts. (f) Statistics of panels a-e. The number (n) of hearts analyzed in each group is indicated in each group. Scale bar, 100μm. |