- Title
-
Spliceosomal protein eftud2 mutation leads to p53-dependent apoptosis in zebrafish neural progenitors
- Authors
- Lei, L., Yan, S.Y., Yang, R., Chen, J.Y., Li, Y., Bu, Y., Chang, N., Zhou, Q., Zhu, X., Li, C.Y., Xiong, J.W.
- Source
- Full text @ Nucleic Acids Res.
|
The CNS and spinal cord undergo apoptosis and fail to differentiate in fn10a mutants. (A, A) An fn10a mutant (A) showing morphological defects in the brain and tail compared with a wild-type sibling (sib) embryo (A). (B, B’) TUNEL staining showing that the fn10a mutant (B) had severe apoptosis in the CNS and neural tube. (C–F, C’–F’) In situ staining for neural markers revealed that differentiated neuronal markers (D–D’, map2; E–E’, tuba; F–F’, pax2) but not a neural progenitor marker (C–C’, nestin) were affected in fn10a mutants (C–F) compared with those in their siblings (C–F). Scale bars, 200 μm. The number of embryos assayed was ≥13 for each panel. |
|
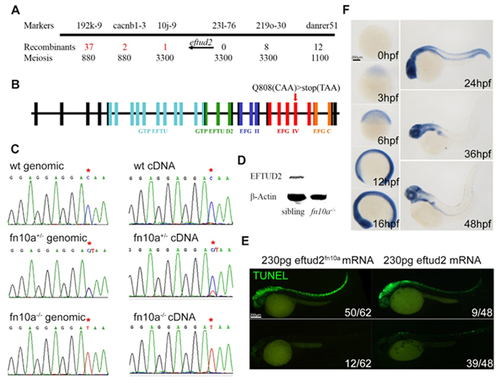
The fn10a locus encodes Eftud2 in zebrafish. (A) Regional fine-map of fn10a with the flanking genetic markers in chromosome 3, showing that eftud2 is located between genetic markers 10j-9 and 231–276. (B) A C-to-T mutation in exon 24 led to premature termination of eftud2 translation that produced mutant Eftud2 protein lacking 165 amino-acids in the C-terminus. (C) Sequencing data of genomic and cDNA of wild-type sibling, heterozygous, and mutant fn10a embryos. Note that mutant eftud2 mRNA is not stable in fn10a+/− mutants. (D) Western blot showing that Eftud2 protein was not detectable in fn10a−/− mutants. β-Actin was used to normalize protein loading. (E) Left panels: TUNEL staining for neuronal apoptosis in only 12/68 fn10a mutants was rescued (lower panel) while that in 50/62 mutants was not rescued (upper panel) by mutant eftud2fn10a mRNA. Right panels: TUNEL staining for neuronal apoptosis in 39/48 fn10a mutants was rescued (lower panel) but that in 9/48 mutants was rescued less (upper panel) by wild-type eftud2 mRNA. All fn10a mutants were confirmed by PCR-based genotyping. (F) Whole-mount in situ hybridization revealed that eftud2 was broadly expressed at 3, 6, 12, 16 and 24 hpf and enriched in the brain and branchial arches at 36 and 48 hpf. Scale bar, 200 μm. |
|
Neuronal progenitors undergo apoptosis in both the head and spinal cord of eftud2fn10a mutants. Immunostaining and TUNEL staining in cryosections of the embryonic brain (A–L, A'–L') and spinal cord (M–T, M'–T') at 36 and 48 hpf. (A–D, A'–D') The neural progenitor marker Sox2 was co-localized with apoptotic (TUNEL+) cells in fn10a mutants (B, B', D, D'). Panels A' to D' are expanded from the boxed areas in panels (A to D). Apoptotic cells were hardly detectable in wild-type siblings (A, A', C, C'). (E–H, E'–H') The postmitotic neuronal marker Huc at 36 hpf was not co-localized (F, F'), but was partially co-localized at 48 hpf (H, H') with apoptotic cells in fn10a mutants. (I–L, I'-L') The mitotic-phase marker pH3 was barely co-localized with apoptotic cells in fn10a mutants at 36 hpf and 48 hpf. Note that the number of pH3+ cells was dramatically higher in fn10a mutants at 48 hpf (L, L') than at 36 hpf (J, J'). (M–N') The neuronal precursor marker sox2 was co-localized with apoptotic cells in the spinal cord in fn10a mutants. Note more TUNEL-positive cells in fn10a mutants (M', N'). (O–P') Immunostaining revealed the disruption of neural-glial cell formation with more TUNEL-positive cells in fn10a mutants (O', P') than in sibling controls (O, P). (Q–R') Newly-generated Huc+ neurons were less affected in fn10a mutants at 36 hpf (Q'), but they underwent apoptosis at 48 hpf (R'). (S–T', U–V') The mitotic-phase marker pH3 was increased in fn10a mutants at 48 hpf and partially overlapped with apoptotic cells (S', T'). (M–T') Transverse views; (U–V') sagittal views. Scale bars in A–D, E–L', 50 μm; A'–D', M–T', 10 μm; U–V', 30 μm; n ≥ 5 for each panel. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
The p53 signaling pathway is activated in fn10a mutants. (A) Gene expression in RPKM (Reads Per Kilobase of transcript per Million mapped reads) in mutants and WT-siblings. Yellow asterisks, p53 pathway genes; red circles, genes of the p53 pathway differentially expressed in mutants and their siblings; green circles, intron-retained genes in the p53 pathway; and blue circles, other RefSeq genes. (B) Genes of the p53 pathway were up-regulated and enriched with intron retention but few had both intron retention and increased mRNA levels in fn10a mutants. (C) Real-time PCR confirmed the transcriptional up-regulation of p53 pathway genes (n = 5). (D) Knockdown of p53 expression in fn10a mutants rescued neuronal apoptosis in the CNS and spinal cord. Note that ∼50% of p53MO-injected fn10a mutants had no TUNEL signals and ∼42% had fewer TUNEL signals. (E) Over-expression of upf1 mRNA partially reduced p53 expression in eftud2 morphants while it had no effect on p53 expression in wild-type embryos (n = 5). |
|
Many organs other than the brain and neural tube were not affected in fn10a mutants. RNA in situ hybridization with the organ-specific markers skeletal muscle myoD (A, A'), macrophage lysozyme C (B, B'), hematopoietic cmyb (C, C'), digestive organ foxa3 (D, D'), kidney pax2 (E, E') and wt1a (F, F'), ventricle myosin heavy chain (vmhc) (G, G'), and atrium myosin heavy chain (amhc) (H, H') in fn10a siblings (A-H) and fn10a mutants (A'-H'). Note that these organ-specific genes appeared to be normally expressed in fn10a mutants compared with those in wild-type siblings. Scale bars, 200 μm. The number of embryos assayed was ≥14 for each panel. |
|
Apoptosis was mainly found in neuronal progenitors in fn10a mutants. (A-A', B-B') Transient expression of the neuronal progenitor reporter Tg(ngn1:GFP) co-localized with TUNEL-positive cells in fn10a mutants (B-B') compared with those in wild-type siblings (A-A') at 36 hpf. (C-C', D-D') Transient expression of the neuronal-glial cell reporter gfap:GFP rarely co-localized with TUNEL-positive cells in fn10a siblings and mutants. Scale bars in A to D, 70 μm; in A' to D', 10 μm. The number of embryos for each assay was ≥5. |
|
Intron-retaining transcripts are enriched in the brain. Whole-mount RNAscope in situ hybridization assays showing the expression pattern of intron-retaining transcripts. (A, A') odc1, as a positive control, was broadly expressed in both siblings (A) and fn10a mutants (A'). (B, B') dapB, as a negative control, had little or no signal, except in the ear and notochord region, in siblings (B) and mutants (B'). (C-C', D-D') Intron-containing probes for ccnb1 and tardbp were enriched in the brain of fn10a mutants (C', D'; arrows) compared with siblings (C, D). Scale bars, 400 μm; n ≥10 for each panel. |
|
Intron-retaining transcripts are located in both nucleus and cytoplasm. RNAscope in situ hybridization assays showing subcellular localization of intron-retaining transcripts on cryosections of fn10a embryos at 36 hpf. DAPI staining (blue) was used to mark the nucleus. (A-A'', B-B'') odc1, as a positive control of RNAscope, was broadly expressed in both siblings (A-A'') and mutants (B-B''). (C-C'', D-D'') dapB, as a negative control of RNAscope, had no signal in siblings (C') and mutants (D'). (E-E'' to H-H'') The intron-containing probes of ccnb1 and tardbp had higher expression in fn10a mutants (F', H') than in siblings (E', G'). Note that retained introns were found in both nucleus and cytoplasm in fn10a mutants although more were found in the nucleus. Arrows indicate their cytoplasmic locations (F'', H''). Scale bars, 10 μm; n ≥5 for each panel. |