- Title
-
RhoA/ROCK pathway activity is essential for the correct localization of the germ plasm mRNAs in zebrafish embryos
- Authors
- Miranda-Rodríguez, J.R., Salas-Vidal, E., Lomelí, H., Zurita, M., Schnabel, D.
- Source
- Full text @ Dev. Biol.
|
Rho GTPases immunofluorescense pattern during germ plasm localization at 8-cell stage zebrafish embryos. (A) RhoA immunolocalization. Arrow indicates first cleavage and arrowhead second cleavage furrows that are positive for RhoA (n=31 of 6 independent experiments). (B) Cdc42, immunolocalization (n=8 of 2 independent experiments). (C) Rac1 immunolocalization (n=7 of 2 independent experiments). Asterisks indicate cleavage furrow. (A, B and C) Nuclei were stained with DAPI and are shown in cyan. (D) RhoA (green), immunolocalization and vasa mRNA (red), localization by fluorescent in situ hybridization (n=9 in 2 independent experiments). (E) 3D reconstruction models of the image shown in D, rotated to highlight the regions indicated by the arrow, the single asterisk, the arrowhead and the double asterisks. The inserts indicated by the discontinuous lined squares are magnified and shown on the right side of each 3D model. Scale bar 250 μm. EXPRESSION / LABELING:
|
|
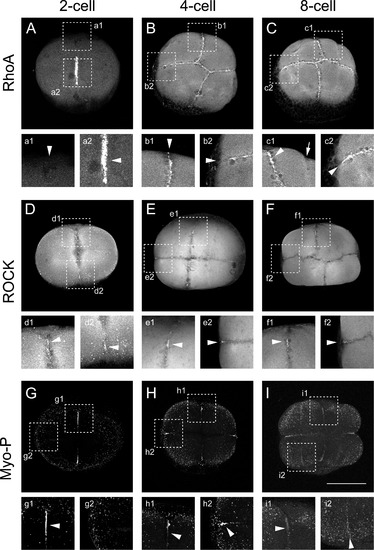
RhoA/ROCK/myosin pathway is localized at the cleavage furrow during the first cell divisions. (A) RhoA immunolocalization at early cleavage in 2-cell (n=16); (B) 4-cell (n=22) and (C) 8-cell embryos (n=9) of four independent experiments. Insets show the immunolocalization signal at the edge of the first cleavage furrow (a1) or the most animal region of the first cleavage furrow in 2-cell embryo shown in A. (b1), first cleavage furrow (b2), second cleavage furrow in 4-cell embryo. (c1), first cleavage furrow and (c2), second cleavage furrow, in 8-cell embryo. (D) ROCK immunolocalization in, 2-cell (n=10); (E) 4-cell (n=18) and (F) 8-cell embryos (n=24) of six independent experiments. Insets show magnification of the first or second cleavage furrow at 2-cell (d1 and d2, respectively), 4-cell (e1 and e2, respectively), 8-cell stage (f1 and f2, respectively). (G)Phosphorylated myosin immmunolocalization in 2-cell (n=5), (H) 4-cell (n=21) and (I) 8-cell embryos (n=5) of five independent experiments. Insets show magnification of the first or second cleavage furrow at 2-cell (g1 and g2, respectively), 4-cell (h1 and h2, respectively), 8-cell stage (i1 and i2, respectively). Scale bar 250 μm. EXPRESSION / LABELING:
|
|
RhoA inhibition interferes with animal germ plasm localization. RhoA immunolocalization (A, B, G and H) vasa mRNA localization (C, and D), nanos mRNA localization (I and J) by fluorescent whole mount in situ hybridization. Embryos injected with buffer (A, C and E; n=9) and (G, I and K; n=8) or with the RhoA inhibitor C3 (B, D and F; n=4) and (H, J and L; n=15). (E and F; K and L) images superimpositions of double labeled embryos with RhoA immunolocalization (green) and vasa or nanos fluorescent in situ hybridization (red) respectively. Insets show image magnification of the corresponding first two-cleavage furrow intersection (e1, f1, k1 and l1) or cleavage furrow edge (e2, f2, k2 and l2). Scale bar 250 μm. |
|
mRNAs of vasa and nanos are mislocalized when RhoA is inhibited with C3. nanos mRNA in situ hybridization (A and B) and corresponding magnification insets (a and b). vasa mRNA in situ hybridization (C and D) and corresponding magnification insets (c and d). Embryos were injected Buffer (A, C, a and c) or with C3 (B, D, b and d) arrows indicate that there is no signal at the end of the cleavage furrow. Asterisk shows the characteristic localization of vasa mRNA at the periphery of the blastodisc present in control embryos (C) and absent in C3 injected embryos (D). (E) Normal localization of nanos of control embryos (n=94) and C3 injected embryos (n=99) were quantified and statistical difference was observed (left panel; 4 independent experiments). Normal localization of vasa observed in control embryos (n=98) and C3 injected embryos (n=113) was quantified (right panel; 5 independent experiments). The error bars represent the standard error of the mean (** p< 0.01 ***p<0.001). |
|
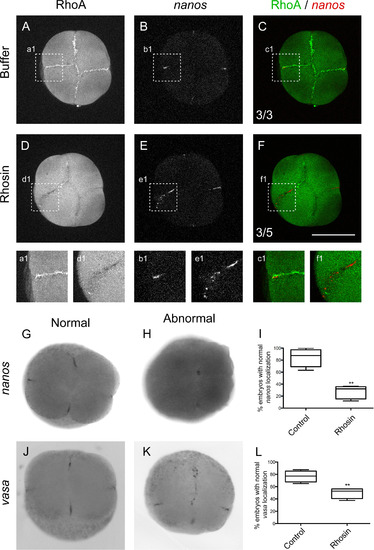
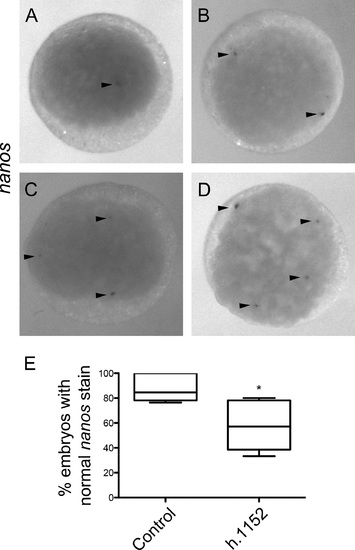
RhoA Inhibition with Rhosin affects vasa and nanos mRNAs localization at the cleavage furrow in 4-cell stage zebrafish embryos. Embryos were injected with control media (A–C, a1, b1, c1, G and J) or with RhoA inhibitor Rhosin (D–F, d1, e1, f1, H and K). (A and D) RhoA immunolocalization. (B and E) nanos mRNA fluorescent in situ hybridization. (C and F) Image superimposition of double labeled embryos with RhoA immunolocalization (green) and nanos fluorescent in situ hybridization (red) Embryos injected with control media show normal nanos localization (n=3); meanwhile Rhosin injected embryos showed abnormal localization (n=5). (G) nanos normal localization and (H)abnormal localization of nanos at the center of the injected embryo with Rhosin. vasa mRNA in situ hybridization normal localization (J) and abnormal vasa localization in injected embryos with Rhosin (K). (I) The differences observed in the pattern of normal localization of nanos in control (n=80) and Rhosin injected embryos (n=97) were quantified in 5 independent experiments. The error bars represent the standard error of the mean (** p<0.01). (L) Differences observed in the normal localization pattern of vasa between control (n=79) and Rhosin injected embryos (n=83) were quantified from 3 independent experiments. The error bars represent the standard error of the mean (** p<0.01). Scale bar 250 µm. |
|
Rhosin injections at 1-cell stage cause significant decrease of primordial germ cells (PGCs) in 24-h zebrafish embryos. (A and E) nanos mRNA in situ hybridization in 24-hpf zebrafish embryos showed the normal staining of the PGCs in embryos at the prospective gonads. Injected embryos with Rhosin showed less nanos staining at only one side of the embryo and signal was not detected at the other side (B and F) Decrease in PGCs is evident in Rhosin treated embryos as detected by nanos mRNA in situ hybridization in embryos (C and G). Ectopic signal of nanos is shown with arrowhead (C and G). Embryos with out positive nanos cells (D and H). (A-D) lateral view and (E-H) dorsal views. (I) The embryos with normal nanos localization of injected control embryos (n=79) and Rhosin injected embryos (n=83) were quantified in 5 independent experiments. The error bars represent the standard error of the mean (** p<0.01). |
|
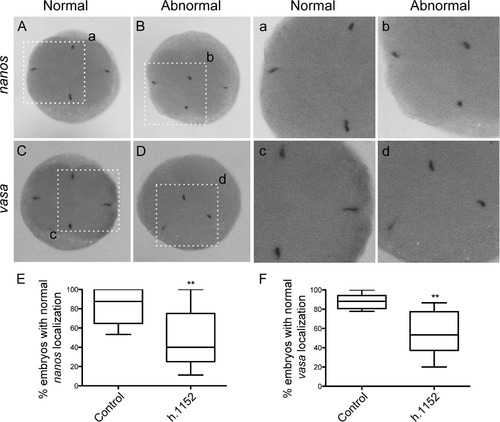
ROCK inhibition disturbs the localization of vasa and nanos mRNAs at the distal part of the cleavage furrow at 4-cells stage zebrafish embryos. nanos and vasa in situ hybridization localization in 4-cell stage embryos with the normal localization at the distal edge of the cleavage furrow (A, a1 and C, c1 respectively). h1152 Injected embryos showed abnormal localization of nanos (B and b1) and vasa (D and d1) at the center of the cleavage furrow. Quantification of normal localization of the mRNAs aggregates of nanos (E) or vasa (F), in embryos injected with control buffer or h1152. (E) Localization of nanos in control (n=160) and embryos injected with h1152 (n=209) showed statistical differences. (F) Localization of vasa in control (n=138) and h1152 treated embryos (n=131) also showed statistical differences from 4 independent experiments. The error bars represent the standard error of the mean (** p<0.01). |
|
Inhibition of ROCK activity generates embryos with less vasa positive cells. Characteristic vasa staining at 24 hpf embryos showing localization at the beginning of the yolk extension in lateral view (A and a) and at the dorsal view two groups of cells mark the prospective gonads at each side of embryo (C and c). Injected embryos with h1152 showed less vasa positive cells (B and b), and at the dorsal view injected embryo with h1152 showed vasa positive cells only at one of the prospective gonads (D and d). Also ectopic signal was observed in injected embryos with h1152 (E). Embryos with the normal vasa localization at 24 hpf were quantified in injected embryos with control buffer (n=85) or in embryos injected with h1152 (n=93) and statistical difference were observed in 3 independent experiments. The error bars represent the standard error of the mean (** p<0.01). |
|
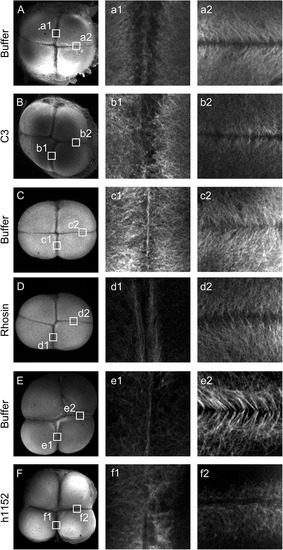
Inhibition of RhoA/ROCK pathway affects remodeling of the furrow microtubule array. Buffer (A, C and E) and C3, Rhosin and h1152 (B, D and F respectively) injected embryos labeled with anti α tubulin antibody. In buffer injected embryos the furrow microtubule array form as a parallel array at the center of the cleavage furrow at mature furrow (A, a1; C, c1 and E, e1) meanwhile in injected embryos with C3 (B, b1), Rhosin (D, d1) or h1152 (F, f1) no deposition of the microtubules at the center of the cleavage furrow was observed. Upon furrow maturation furrow microtubules appear at the distal edge in a characteristic V shape in buffer injected embryos (A, a2; C, c2 and E, e2). Microtubules from injected embryos with C3, Rhosin or h1152 did not showed this characteristic pattern (B, b2; D, d2 and F, f2 respectively) but maintained a perpendicular arrangement towards the cleavage furrow (b2 and d2) or showed a complete lack of organization (f2). Data was analyzed from 3 independent experiments of C3 injected embryos (n=11) or buffer injected embryos as control (n=7); from 4 independent experiments of Rhosin injected embryos (n=12) or buffer injected embryos as control (n=15) and from 4 independent experiments of h1152 injected embryos (n=13) or buffer injected as control (n=11). |
|
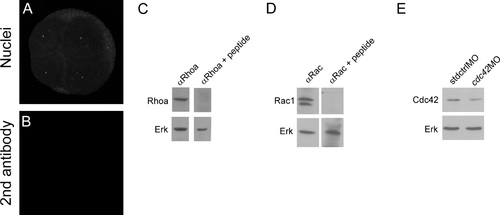
Rhoa, Cdc42 and Rac1 antibodies recognize their respective proteins. Embryos at 4 to 8- cell transition were co-stained with DAPI to show the nuclei (A) and does not show any positive signal after inmunodetection with only anti-rabbit secondary antibody (B). Whole protein samples were obtained from 24 hpf embryos. A single band of the expected size is observed in the western blot against RhoA (C top left panel). Incubation of the primary RhoA antibody with RhoA specific peptide competes and the band in the western blot is not longer observed (C top right panel). Membranes were stripped and blotted against Erk as loading control (C bottom left and right panels). A doublet is observed in western blot against Rac1 (D top left panel) that was competed with specific peptide against Rac1 (D top right panel), membranes were stripped and blotted against Erk as loading control (D bottom left and right panel). Cdc42 specificity was demonstrated by using specific morpholino against cdc42 (cdc42MO). The amount of Cdc42 is less in cdc42 morfants compared to embryos injected with standard morpholino (stdMO). Membranes were stripped and blotted against Erk as a loading control. Western blots experiments were repeated at least in 6 times in independent experiments and antibodies validation were repeated 3 times. |
|
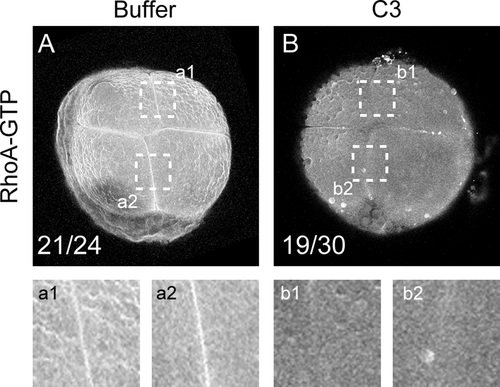
C3 affects the localization of active RhoA at the cleavage furrow. (A and B) Active RhoA was immunolocalized with an anti-active-RhoA (RhoA-GTP) mouse monoclonal antibody and visualized by confocal microscopy. (A) Embryos were injected with control buffer (n=24) or with (B) C3 (n=30). a1 and a2 show magnification of regions highlighted in A. b1 and b2 show magnification of regions highlighted in B. |
|
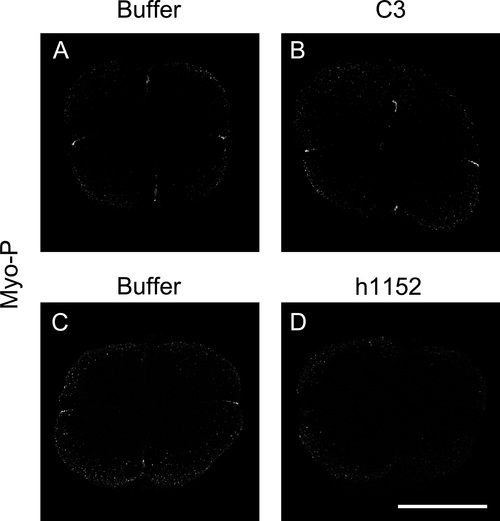
Myosin Light Chain phosphorylation is affected when RhoA/ROCK pathway is inhibited. Embryos were injected either with RhoA inhibitor (C3) or ROCK inhibitor (h1152) and as control embryos were injected with buffer. At 4-cell stage inmunodetection for myosin light chain phosphorylation (MyoP) was performed. (A and B) Inhibition of RhoA with C3 (n=24) showed the distribution of MyoP at the center of the embryo and diminished compared to buffer (n=11) injected embryos of 2 independent experiments. (C and D) Inmunodetection of MyoP of embryos injected with h1152 (n=41) showed less positive signal compared to buffer (n=32) injected embryos of four independent experiments. |
|
Inactivation of ROCK affects PGCs specification. (A, B, C and D) nanos mRNA in situ hybridization localization in 128 to 256-cell stage embryos. Arrowheads indicate the positive signal. Embryos at 128–256 cell stage normally have four cells that express mRNAs of the GP such as nanos (D), injected embryos with h1152 showed less positive cells as shown in (A, B and C). (E) The number of embryos with a normal number of positive cells for nanos was quantified in 128–256 cells embryos injected at one cell stage, either with control buffer (n=46) or with h1152 (n=63). The error bars represent the standard error of the mean (* p<0.05). |
|
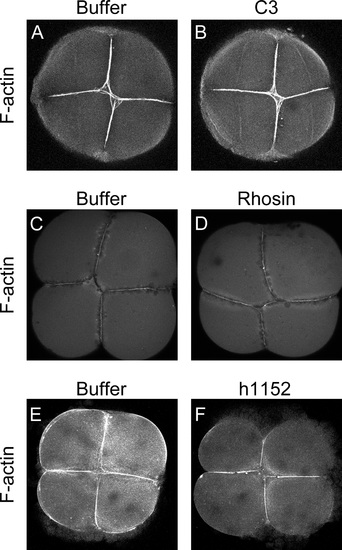
Inhibition of RhoA/ROCK pathway does not affects actin filaments localization at the cleave furrow. (A and B) 4-cell embryos stained with phalloidin after injection with either buffer (n=35) or C3 (n=52) showed no difference in 6 independent experiments. (C and D) 4-cell embryos stained with phalloidin after Rhosin injection (n=25) (D) or Buffer injection (n=27) (C) showed no differences between them in 5 independent experiments. (E and F) Embryos injected either with buffer (n=5) or h1152 (n=7) showed no differences when stained with phalloidin to detect filamentous actin at the cleavage furrow in 3 independent experiments. |
|
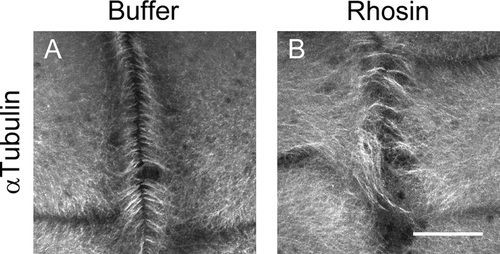
Inhibition of RhoA affects remodeling of the furrow microtubule array. Embryos were injected with RhoA inhibitor (Rhosin) at one cell stage and at 8-cell inmunolocalization of α-tubulin was performed. (A and B) At 8 cell embryonic stage microtubules from injected embryos with Rhosin are disorganized at the cleavage furrow (n=6) compared to the buffer injected embryos (n=6) where the microtubules showed a perpendicular distribution towards the cleavage furrow in 3 independent experiments. |
Reprinted from Developmental Biology, 421(1), Miranda-Rodríguez, J.R., Salas-Vidal, E., Lomelí, H., Zurita, M., Schnabel, D., RhoA/ROCK pathway activity is essential for the correct localization of the germ plasm mRNAs in zebrafish embryos, 27-42, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.