- Title
-
Syndecan-4 modulates the proliferation of neural cells and the formation of CaP axons during zebrafish embryonic neurogenesis
- Authors
- Luo, N., Li, H., Xiang, B., Qiao, L., He, J., Ji, Y., Liu, Y., Li, S., Lu, R., Li, Y., Meng, W., Wu, Y., Xu, H., Mo, X.
- Source
- Full text @ Sci. Rep.
|
Syn4 expression pattern analysis in zebrafish. (A) sqRT-PCR reveals that syn4 is expressed throughout embryonic developmental stages. (B–M) WISH shows the spatiotemporal expression pattern of syn4. (B–D) Syn4 is ubiquitously distributed before the gastrulation. (E) In 75% epiboly, syn4 is expressed in the thin evacuation zone and prechordal plate. (F–K) Syn4 is expressed in ventral diencephalon, midbrain, hind brain and neural crest during Segmentation Period. (L,M) In 48 hpf, syn4 is restricted to the ventricular zone in the forebrain, posterior midbrain and hindbrain. (N) Negative control of syn4 at 48 hpf. EZ: thin evacuation zone, FV: ventricular zone in the forebrain, Hb: hindbrain, Mb: midbrain, MHB: mid-hindbrain boundary, pp: prechordal plate, PM: posterior midbrain, R: rhombomeres, som: somites, sc: spinal cord, vd: ventral diencephalon. Lateral view, dorsal to the right in (B–E), dorsal view, anterior to the left in (G,I,K,M) lateral view, dorsal to the right and anterior to the top in (F), lateral view, dorsal to the top and anterior to the left in (H,J,M). EXPRESSION / LABELING:
|
|
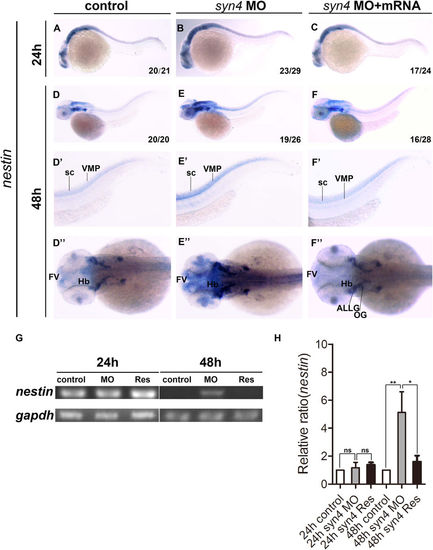
Knockdown of syn4 expression up-regulated the expression of nestin. (A–C) Expression of nestin at 24 hpf, lateral view. (D–F) Expression of nestin at 48 hpf, lateral view. (E–E”) Nestin transcript level was increased significantly in syn4 morphants, dorsal view. (F–F”) The defective phenotypes were rescued by co-injecting syn4 mRNA. (G) sqRT-PCR showed the similar results. (H) Densitometric quantification of sqRT-PCR. The relative nestin expressing level is significantly higher in syn4 morphants (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student’s unpaired t-test). ALLG: anterior lateral line ganglion, FV: ventricular zone in forebrain, Hb: hindbrain, OG: octaval ganglion, sc: spinal cord, VMP: ventral motoneuron precursors. |
|
Loss of syn4 increased neuroglial cells. (A–R'') WISH results of gfap (A–F''), slc1a3a (G–L'') and olig2 (M–R'') at 24 hpf and 48 hpf in zebrafish. (D–F') The expression of gfap, (J–L'') slc1a3a, (Q–R'') olig2 are higher in the syn4 morphants than controls at 48 hpf. (S) sqRT-PCR analyzes of the transcript level of gfap, slc1a3a and olig2. (T–W) Densitometric quantification of sqRT-PCR. The syn4 morphants shown a higher expression level of gfap, slc1a3a and olig2 (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student's unpaired t-test). FV: ventricular zone in forebrain, Hb: hindbrain, PM: posterior midbrain, sc: spinal cord. Lateral views, dorsal to the top and anterior to the left in (A–C,D–F,G–I,J–L',M–O,P–R,P''–R''), Dorsal view, anterior to the left in (D'–F',G'–I',J''–L'',P'–R'). |
|
Knockdown of syn4 expression promotes the generation of neurons. (A–R'') WISH results of elavl3, islet1, and islet2a at 24 hpf and 48 hpf in zebrafish. (E–E'') The expression of elavl3, (K–K'') islet1, (Q–Q'') islet2a in spinal cord were increased in syn4 morphants at 48 hpf. co-injecting syn4 mRNA was able to rescue the defects. (S) sqRT-PCR analyzes of the transcript level of elavl3, islet1 and islet2a. (T–V) Densitometric quantification of sqRT-PCR. The syn4 morphants shown a higher expression level of elavl3, islet1 and islet2a (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student's unpaired t-test). ALLG: anterior lateral line ganglion, Hb: hindbrain, Mb: midbrain, sc: spinal cord. Lateral views, dorsal to the top and anterior to the left in (A–F',G–L',M–R'). Dorsal view, anterior to the left in (D''–F'',J''–L'',P''–R''). |
|
Whole-mount immunohistochemistry staining of PCNA and H3S10 in zebrafish embryos. (A–D'') Immunostaining of PCNA and H3S10 in 24 hpf and 48 hpf embryos, the box regions are magnified. (E,F) Quantification of PCNA-positive cells (E) and H3S10 -positive cells (F) in different treatment groups. Both PCNA positive cells and H3S10 positive cells are considerably increased in syn4 morphants and the defects were partially rescued by co-injection with syn4 mRNA. For each group, 18 embryos are scored. (mean ± s.e.m, n = 3, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, Student's unpaired t-test). (G) The double WISH with H3S10 and olig2. Arrows label the H3S10 positive cells, the box regions are magnified. Lateral views, dorsal to the top and anterior to the left in (A–D'',G). |
|
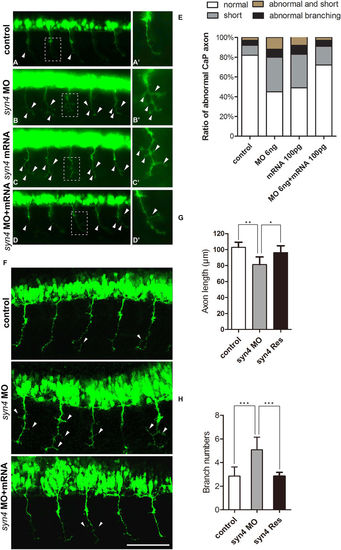
Syn4 is required for the CaP axons formation. (A–D') Embryos at 26 hpf show PMNs in green fluorescence. (B,B') Syn4 morphants show shortened and branching CaP axons, (C,C') syn4 mRNA-injected embryos have same phenotypes as syn4 morphants. (D,D') Co-injection of syn4 MO with syn4 mRNA partially rescues CaP axon defects. (E) Statistical results of CaP axon outgrowth are shown in (A–D'). For each group 60 axons from 12 embryos are scored. (F) The CaP axons of control embryos and syn4 morphants are photographed under confocal microscopy. (G) Statistical results of axon length in (F–G). The length of CaP axons is reduced in syn4 morphants (**P < 0.01, t-test). (H) the branch number of CaP axons is increased (***P < 0.001, t-test) in syn4 morphants. For each group 75 axons from 15 embryos are scored. The box regions are magnified and show in (A'–D'). Arrows indicate shortened CaP axons and abnormal branchs. Lateral views, dorsal to the top and anterior to the left in (A–D’,F). Scale bars: 100 μm. |
|
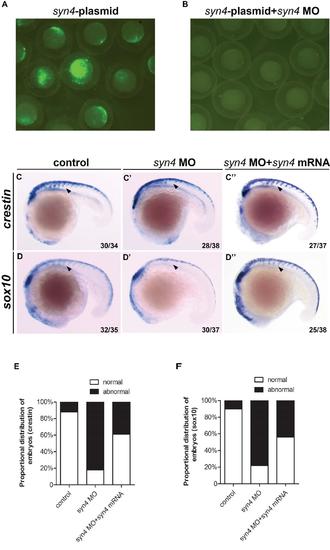
Syn4 is required for the migration of neural crest. (A, B): Knockdown efficiency of syn4 MO at 9 hpf. (A): Zebrafish embryos were injected with 30 pg of pEGFP-N1-syn4 recombinant plasmid containing the MO-binding site. (B): Green fluorescence was totally abolished by co-injection with 6 ng syn4 MO. (C-D''): WISH assays of crestin (C-C''), sox10 (D-D'') at 18 somites stage. Arrows indicate the expression of crestin/sox10 in the morphants are inhibited. (E, F): Quantification of phenotypes produced by syn4morphants at 18 somites stage, with or without syn4 mRNA. Lateral views, dorsal to the top and anterior to the left in C-D’’. |
|
Knockdown syn4 disrupt the muscle patterning of zebrafish. In situ hybridization for indicated genes.(A-D'') WISH assays of myod at 18 somites stage,white bars marks vertical. The morphants and syn4-C2 mRNA injected fishs appeared abnormal curvature. (E):Quantification of phenotypes at 18 somites stage.(F-K): Expression pattern of islet1/islet2a in different embryos at 18 somites stage. |
|
Knockdown syn4 disrupt the muscle patterning of zaebrafish. (A-C'') Fast and slow muscle structuresinTg(Hb9:GFP line) at 18s. Fast and slow muscles in the trunk are stained with phalloidin, which recognizes actin fibers of both muscle types, PMNs in green fluorescence. Phalloidin labeling revealed an impaired muscle structure in morphants.(D-F'') muscle structures inTg(Hb9:GFP line) at 26 dpf. Phalloidin labeling revealed the fibers are abnormally thick and wavy in morphants. The box regions were magnified. |
|
The C2 domain of syn4 is crucial for the proliferation of neural cells and the formation of CaP axons (A): Schematic representation of Syndecan 4 constructs. (B-G''): Expression pattern of nestin in different embroyos at 24 hpf (B-D) and 48 hpf (E-G''). (H): Embryos at 26 hpf showing PMNs in green fluorescence. The box regions were magnified. Arrows indicate shortened CaP axons and abnormal branchs. (I): Statistical results of relative ratio of nestin expression level in (E-G'').(J): Quantification of CaP axon branching in(H),the number of axonal branches was significantly higher in syn4 morphants and syn4-ΔC2 mRNA injected embryos than in controls.For each group 84 axons from 14 embryos were scored(mean ± s.e.m,n=3, ***P<0.001, **P<0.01, *P<0.05, ns= not significant,t-test).Hb: hindbrain, sc: spinal cord. |
|
Overexpression of syn2 is unable to rescue the phenotypes caused by knockdown of Syn4 functions In situ hybridization for indicated genes. (A-F'): WISH assays of gfap and elavl3 in different embryos at 48 hpf. The higher expression level of gfap/elavl3 in morphants at 48 hpf couldn’t be rescued by co-injection with syn2 mRNA. FV: ventricular zone in the forebrain, Hb: hindbrain, Mb: midbrain. |
|
Immunofluorescence staining of H3S10 in zebrafish embryos (A-C'): Immunostaining of H3S10 in 48 hpf embryos. (D-F''): Transverse sections of forebrain (Top), midbrain (middle) and hindbrain (below) of different embryos immunostained for H3S10 (green).G: Quantification of H3S10 -positive cells. For each group, 6 embryos are scored (mean ± s.e.m,n=3, ***P<0.001, **P<0.01, *P<0.05, ns= not significant, Student’s unpaired t-test). e: eye, Fb: forebrain, Hb: hindbrain, Mb: midbrain,t: tectum.The dotted boxes mark the brain regions. |