- Title
-
Transgenic FingRs for Live Mapping of Synaptic Dynamics in Genetically-Defined Neurons
- Authors
- Son, J.H., Keefe, M.D., Stevenson, T.J., Barrios, J.P., Anjewierden, S., Newton, J.B., Douglass, A.D., Bonkowsky, J.L.
- Source
- Full text @ Sci. Rep.
|
FingR constructs and testing in zebrafish. (A) Schematic diagram of FingR constructs used to generate plasmids and transgenic lines. FingR(PSD95)-GFP binds endogenous PSD-95 protein at the post-synaptic density; FingR(GPHN)-Cherry binds endogenous GPHN. (B) Use of inducible Gal4/UAS system with FingR(GPHN) or FingR(PSD95) in different Gal4 lines for differential synapse labeling (drawn by JHS). (C) Contrasting examples of unregulated (FingR-GFP) and regulated (FingR(PSD95)-GFP) FingR plasmids injected into Tg(otpb.A.Gal4) embryos. GFP signal from the unregulated FingR is distributed throughout the neuron and neurites (arrows) and individual synaptic puncta are not visualizable. In contrast, distinct puncta are seen (open arrowheads) using regulated FingR expression. Confocal images, scale bar 50 μm, 10 μm in inset panels; confocal z-stacks, ventral views, rostral to the top. D) Demonstration of puncta labeling with regulated FingR for GPHN. Confocal z-stacks, ventral view, rostral to top, of Tg(otpb.A:Gal4) embryo injected with FingR(GPHN)-Cherry. Arrows, neuron soma; arrowheads, puncta. Scale bar 50 μm, 10 μm in inset panel. |
|
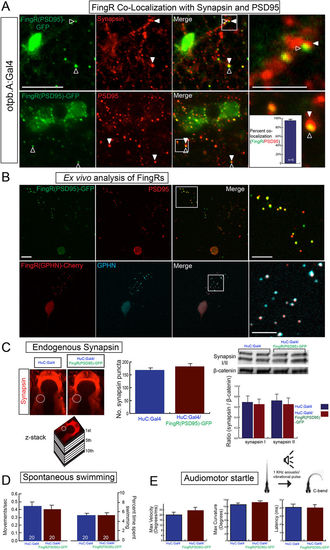
Transgenic FingR lines reflect endogenous synapses and do not affect synapse number or behavior. (A) FingR(PSD95)-GFP signal is adjacent to Synapsin immunohistochemistry (top panels) and overlaps PSD-95 immunohistochemistry (bottom panels). Quantification of FingR(PSD95)-GFP signal and endogenous PSD-95 signal demonstrates 95% overlap of PSD-95 immunohistochemistry with GFP signal from FingR (bar graph: median 95% +/− 1%, SEM; n = 6 larvae; p 0.01). Confocal images of sections from immunostained Tg(otpb.A:Gal4); Tg(FingR(PSD95)-GFP) larvae, scale bar 10 μm, 5 μm in inset panels. (B) Ex vivo sparse zebrafish primary neuron cell culture demonstrates co-localization of FingRs and endogenous synaptic proteins. Top row, dissociated neurons from Tg(otpb.A:Gal4); Tg(FingR(PSD95)-GFP) embryo, immunohistochemistry for anti-GFP and anti-PSD95. Bottom row from Tg(HuC:Gal4); Tg(FingR(GPHN)-Cherry) embryo, immunohistochemistry for anti-mCherry and anti-GPHN. Confocal images of slides, scale bar 10 μm, 5 μm in inset. (C–E) Pan-neuronal expression in Tg(FingR(PSD95)-GFP); Tg(HuC:Gal4) larvae. (C) FingR(PSD95)-GFP expression does not affect Synapsin protein expression. Data is double-transgenic larvae compared to Tg(HuC:Gal4) only. Images are confocal z-stacks, dorsal views, rostral to top; dotted circle indicates area of Synapsin puncta quantification for bar graphs. Western blots are for Synapsin in whole larvae, standardized to β-catenin; quantified bar graphs to right. (D) FingR(PSD95)-GFP expression does not affect spontaneous swimming behavior (percent time swimming and number of movements), n = 20 embryos each test; three separate experiments; SEM. (E) FingR(PSD95)-GFP expression does not affect audiomotor startle response, n = 27 (control) and 30 (FingR) embryos each test; three separate experiments, SEM (velocity, body curvature, or latency). |
|
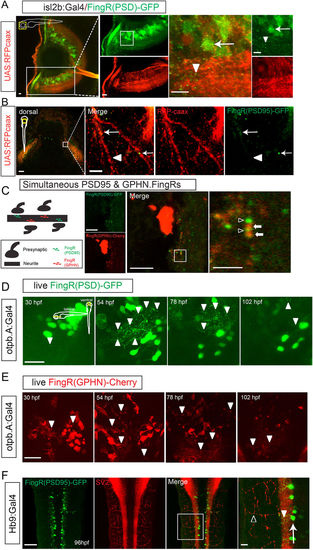
Use of FingRs in visualizing and tracking synapses. (A) Lateral view of eye from transgenic zebrafish expressing FingR(PSD95)-GFP and RFP-caax in retinal ganglion neurons (transgenic Tg(isl2b:Gal4); Tg(UAS:RFP-caax); Tg(FingR(PSD95)-GFP)). GFP expression is seen in puncta (arrowheads) and neuron somas (arrow); RFP labels neurons, dendrites, and their axons as they project towards optic chiasm (Supplemental Movie 1). Confocal z-stacks at 72 hpf, immunohistochemistry for GFP and RFP, scale bar 10 μm. (B) Dorsal view of tectum in Tg(isl2b:Gal4); Tg(UAS:RFP-caax); Tg(FingR(PSD95)-GFP) embryo. Arrow shows FingR(PSD95)-GFP puncta along RGC axon, but not along entire length of axon (arrowhead). Confocal z-stack at 72 hpf, immunohistochemistry for GFP and RFP; scale bar 50 μm, 5 μm in inset panel. (C) Larvae co-expressing FingR(PDS95)-GFP (open arrowheads) and FingR(GPHN)-Cherry (arrows) demonstrates presence of neighboring excitatory and inhibitory puncta. Confocal z-stacks; scale bar 10 μm, 2.5 μm in inset panel. (D,E) FingRs can be used for in vivo visualization and monitoring of synapses. (D) Time-series of dopaminergic neurons and synapses in Tg(otpb.A:Gal4); Tg(FingR(PSD95)-GFP) animal, demonstrating changes in synapse number and expression with development. Scale bar 20 μm. (E) Time-series of dopaminergic neurons and synapses development in Tg(otpb.A:Gal4); Tg(FingR(GPHN)-Cherry) animal, demonstrating changes in synapse number and expression with development. Scale bar 20 μm. (F) Dorsal view in trunk at 96 hpf showing that FingR(PSD95)-GFP is expressed in the spinal cord (arrowhead), but not in axon projections labeled with SV2 (open arrowhead) in the neuromuscular synapses when driven in motor neurons (arrow) by Hb9:Gal4. Confocal z-stacks at 96 hpf, dorsal views, immunohistochemistry for GFP and SV2; scale bar 50 μm, 10 μm in inset panel. |
|
FingRs reveal impairment of synapse development by developmental hypoxic injury. (A) Schematic illustration of hypoxia exposure and imaging. (B) Demonstration of live imaging of effects of hypoxic injury on motor neuron synaptic puncta visualized by FingR(PSD)-GFP in trunk of Tg(Hb9:Gal4); Tg(FingR(PSD95)-GFP) animals. Following hypoxia there is a decrease in the number of puncta labeled by FingR(PSD95)-GFP. Confocal images, rostral to left, scale bar 10 μm. (C) Confocal images and quantification in the dopaminergic neurons in the diencephalon of Tg(otpb.A:Gal4); Tg(FingR(PSD95)-GFP) animals. Hypoxia from 24–48 hpf decreased number of puncta but later hypoxia exposure did not significantly change number; (p = 0.002; one-way ANOVA; SEM shown). Confocal z-stacks, rostral to top, scale bar 10 μm. |