- Title
-
Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration
- Authors
- Beauchemin, M., Smith, A., Yin, V.P.
- Source
- Full text @ Development
|
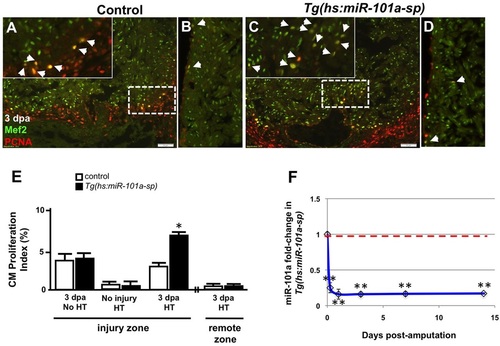
Depletion of miR-101a expression promotes cardiomyocyte proliferation in the injured apex. (A-D) Control or Tg(hs:miR-101a-sp) animals were injured and subjected to daily heat treatment. Hearts were collected at 3dpa for histology, cryosectioned at 10µm and stained with antibodies to detect Mef2 (green) and Pcna (red) to identify proliferating cardiomyocytes (CMs) (arrowheads) at the injured apex (A,C) and lateral wall (remote zone) (B,D). Insets in A,C are higher magnifications of the areas within dashed boxes. (E) CM proliferation indices were determined by representing Mef+Pcna+ cells as a percentage of total Mef2+ cells. Depletion of miR-101a expression doubled CM proliferation indices at 3dpa in the injured apex but not in the remote zone. (F) qPCR studies show ~80% reduction of miR-101a expression levels in Tg(hs:miR-101a-sp) ventricles (blue line) compared with the control group (red dashed line) under conditions of heat treatment. n=5-7; *P<0.05 (Student′s t-test); error bars represent s.e.m. HT, heat treatment. |
|
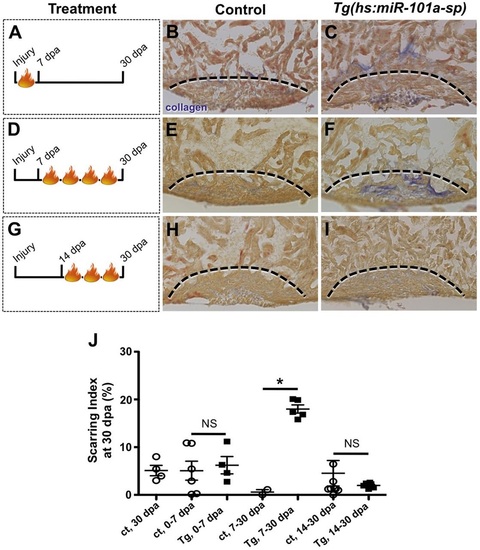
Sustained suppression of miR-101a leads to increased scar tissue. (A-F) Wild-type and Tg(hs:miR-101a-sp) hearts were resectioned, heat treated daily and collected at 7, 14 and 30dpa for histology. Hearts were sectioned at 10µm and stained with Acid Fucshin Orange G (AFOG) to detect muscle (brown), collagen (blue) and fibrin (red). Arrowheads in F mark newly regenerated muscle; dashed lines indicate approximate resection injury plane. The penetrance of each phenotype is indicated (number showing the illustrated phenotype/total number of samples). (G) Scarring indices were established by quantifying the percentage of collagen and fibrin within the total injury area at the indicated time points using CellProfiler. Prolonged depletion of miR-101a led to greater scar tissue at 14 and 30dpa compared with control. (H) Injury size was comparable between the groups as shown by quantification of total injury area. n=5-14; *P<0.001 (Student′s t-test); error bars represent s.e.m. PHENOTYPE:
|
|
Upregulation of miR-101a between 7 and 14dpa is essential for scar tissue removal. (A-I) Control and Tg(hs:miR-101a-sp) animals were subjected to ventricular resection and heat treated at defined intervals. Hearts were collected at 30dpa and processed for AFOG staining to identify scar tissue. (A-C) Heat-treated induced depletion of miR-101a from 0 to 7dpa did not alter scar tissue removal between control and Tg(hs:miR-101a-sp) hearts. (D-F) Suppression of miR-101a expression between 7 and 30dpa resulted in increased scarring in Tg(hs:miR-101a-sp) hearts, compared with controls. (G-I) miR-101a suppression from 14 to 30dpa, however, led to normal scar tissue clearance in control and Tg(hs:miR-101a-sp) hearts. Dashed lines indicate approximate resection injury plane. (J) Quantification of scarring indices demonstrates that increased miR-101a expression at 7-14dpa is required for scar tissue clearance. ct, control. n=4-8; *P<0.01 (Student′s t-test); NS, not significant; error bars represent s.e.m. PHENOTYPE:
|
|
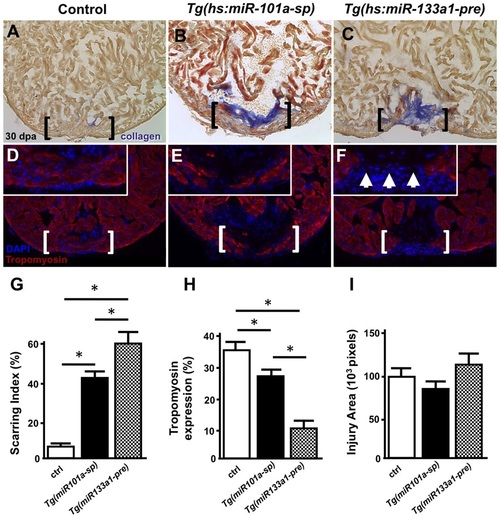
Long-term miR-101a depletion results in muscle regeneration and defects in scar tissue clearance. (A-F) Control, Tg(hs:miR-101a-sp) and Tg(hs:miR-133a1-pre) hearts were resectioned, heat treated daily for 30dpa and extracted for histology. Hearts were cryosectioned at 10µm and stained with either AFOG (A-C) or antibodies directed against Tropomyosin (D-F). Compared with heat-treated control animals, Tg(hs:miR-101a-sp) hearts reveal more scar tissue and less new muscle regeneration in the wounded apex. Tg(hs:miR-133a1-pre) hearts, which overexpress miR-133a1, show defects in both new muscle regeneration and scar tissue removal. Brackets represent approximate injury area; arrowheads in F mark gaps in myocardium and the lack of muscle regeneration. (G-I) Quantification of scarring indices, Tropomyosin expression and injury area were performed with CellProfiler. n=6-7; *P<0.05 (Student′s t-test); error bars represent s.e.m. PHENOTYPE:
|
|
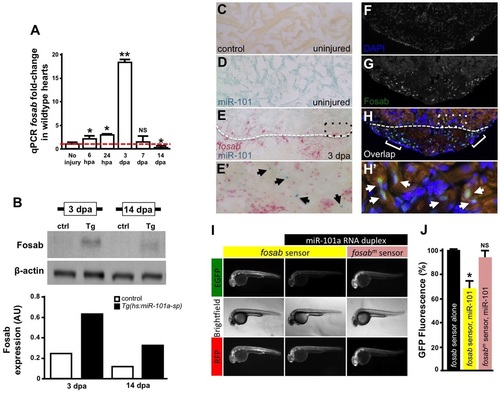
miR-101a controls fosab activity through interactions at the 3′-UTR. (A) qRT-PCR studies show dynamic fosab expression during a time course of normal heart regeneration. (B) Western blots show increased Fosab protein expression in Tg(hs:miR-101a-sp) ventricles at 3 and 14dpa relative to controls. Graph shows quantification of western blots using ImageJ software and normalized to β-actin. (C-E′) RNA in situ hybridization studies to detect miR-101a (blue) and fosab (red) in uninjured and 3dpa regenerating hearts. miR-101a expression is restricted to CMs in uninjured hearts, whereas fosab expression is enriched in CMs at the border zone and non-muscle cells of the wound environment at 3dpa. (F-H′) Antibody staining to detect Fosab protein expression confirms RNA in situ hybridization expression patterns. E′ and H′ are magnified images of boxed areas in E and H; arrowheads mark miR-101a (E′) and Fosab (H′) expression in CMs; brackets in H2 indicate Fosab in the microenvironment. (I,J) mRNA EGFP-fosab-3′-UTR sensor assays reveal ~30% decrease in fosab expression when the sensor is co-injected with miR-101a RNA duplex, relative to EGFP-fosab-3′-UTR sensor injection alone. This miR-101a regulation of fosab is abolished in response to a three-nucleotide mutation of the predicted miR-101a binding site (fosabm sensor). n=6-8 embryos per treatment; *P<0.05, **P<0.001 (Student′s t-test); NS, not significant; error bars represent s.e.m. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Activation of Tg(hs:miR-101a-sp) and Tg(hs:miR-101a-pre) did not induce ectopicprogrammed cell death. Control, Tg(hs:miR-101a-sp) and Tg(hs:miR-101a-pre) hearts were injured, heat-treated and extracted at 3 dpa for analysis of programmed cell death by TUNEL staining. We observed no differences in TUNEL+ cells between control and transgenic animals. An uninjured wildtype heart treated with DNaseI served as a positive control. |
|
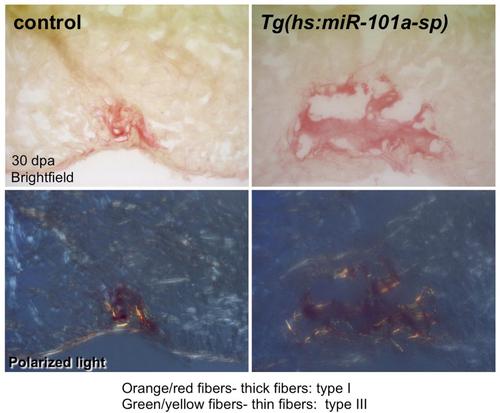
Sustained depletion of miR-101a activity did not alter scar tissue composition. Control and Tg(hs:miR-101a-sp) hearts were resected, subjected to daily heat-treatment and processed for Sirius Red staining. Collagen composition in both control and Tg(hs:miR-101a-sp) hearts is primarily thick fibers (collagen type I, orange). |